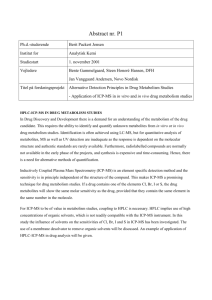
Table 1: ICP-MS and laser conditions
advertisement

Supplementary Material (ESI) for Chemical Communications This journal is © The Royal Society of Chemistry 2002 Laser ablation and ICP-MS operating conditions The gels were ablated using the laser and the resulting plasma analysed using the Platform ICP to give a scanning trace of total isotope concentration (TIC) as a function of distance. The Platform ICP utilises a hexapole as the collision/reaction cell, and so has the capabilities of eliminating Ar based molecular interferences. The Platform ICP-MS conditions used are listed in Table 1. Table 1: ICP-MS and laser conditions. ICP-MS RF Power 1350 W ICP-MS Cool Gas Flow 13.27 L·min-1 ICP-MS Auxilliary Gas Flow 0.82 L·min-1 ICP-MS Nebuliser Gas Flow 1.09 L·min-1 ICP-MS Helium Hexapole Gas Flow 10 mL·min-1 ICP-MS Hydrogen Hexapole Gas Flow 10 mL·min-1 ICP-MS Cone Voltage 70 V ICP-MS Hex Exit Lens Voltage 400 V ICP-MS Hex Bias Voltage 0.1 V ICP-MS Ion Energy 2.0 V ICP-MS Multiplier Voltage 400 V Laser Frequency 200 Hz Laser Power 100 % Laser Spot Size 50 m Laser Scan Speed 20 m·s-1 Supplementary Material (ESI) for Chemical Communications This journal is © The Royal Society of Chemistry 2002 QTOF2 operating conditions The mass spectrometer was operated in the positive ion mode with a source temperature of 80 oC, a counter current gas flow rate of 40 l·h-1 and with a potential of 2600 V applied to the Nanospray continuous LC probe. All data were acquired with the mass spectrometer operating in an automatic data dependent switching mode. The instrument was calibrated with a fourth order calibration using selected ions from Glu-fibrinopeptide-B. Peptide separation protocol The in-gel protein sample was automatically transferred to a 96-well plate and dissected into 1 mm cubes, using the MassPrep station. The gel pieces were destained by means of alternate ammonium bicarbonate and acetonitrile washes. The proteins were then reduced and alkylated with the addition of dithiolthrionine and iodoacetamide. Protein digestion was achieved with the addition of 25 l trypsin solution (Promega, USA) at 6 ng·l-1 for 5 hours at 37 oC. The resulting peptides were extracted in an aqueous solution containing 1 % formic acid and 2 % acetonitrile. The peptides were lyophilised and reconstituted in 5 l of 1 % formic acid. The peptides were separated by means of a Micromass modular capillary LC system connected directly to the Z-spray source of a Micromass Q-TOF2 mass spectrometer. Each sample was loaded on to a C18 pre-column (5 mm length, 320 m ID) at a flow rate of 30 l·min-1 and desalted for 3 min with a solution of 0.1 % formic acid. After desalting on the precolumn, the peptides were directed onto a C18 Picofrit column (5 cm length, 75 m ID), and elution carried out using a solvent gradient starting with 95 % solution A (95 % water, 5 % acetonitrile, 0.1 % formic acid) and 5 % solution B (5 % water, 95 % acetonitrile and 0.1 % formic acid). Following isocratic washing with this buffer, the peptides were eluted using a stepped gradient to 80 % solution B. Sequencing maps for OmpA Supplementary Material (ESI) for Chemical Communications This journal is © The Royal Society of Chemistry 2002 The masses and sequences peptide fragments produced by the trypsin digest of the unknown protein sample were matched with the sequence of OmpA using ProteinLynxTM Global server. Table 2: Analysis of the peptides extracted after the in-gel trypsin digest of the platinum-bound protein, showing (a) the results of the MS/MS sequencing of the peptides produced from the trypsin digest and (b) the peptide fingerprint (bold) mapped onto the whole sequence of the protein. (a) m/z 898.396 Charge 2 MW 1794.751 827.934 705.348 607.828 2 2 2 1653.825 1653.825 1213.612 528.257 458.282 436.777 409.734 2 2 2 2 1054.472 914.518 871.513 817.427 Sequence (R)GMGESNPVTGNTCDNVK(Q) – Carbamidomethylated (K)LGYPITDDLDIYTR(L) (K)LGYPITDDLDIYTR(L) (R)AALIDCLAPDR(R) – Carbamidomethylated (K)DNTWYTGAK(L) (K)AQGVQLTAK(L) (R)RVEIEVK(G) (R)LGGMVWR(A) Residues 279-295 84-97 244-256 298-308 5-13 75-83 309-315 98-104 (b) 1 10 20 30 40 50 60 | | | | | | | AAPKDNTMY TGAKLGWSQY HDTGFINNNG PTHENQLGAG AFGGYQVNPY VGFEMGYDWL GRMPYKGSVE 70 80 90 100 110 120 130 | | | | | | | NGAYKAQGVQ LTAKLGYPIT DDLDIYTRLG GMVWRADTKS NVYGKNHDTG VSPVFAGGVE YAITPEIATR 140 150 160 170 180 190 200 | | | | | | | LEYQWTNNIG DAHTIGTRPD NGMLSLGVSY RFGQGEAAPV VAPAPAPAPE VQTKHFTLKS DVLFNFNKAT 210 220 230 240 250 260 270 | | | | | | | LKPEGQAALD QLYSQLSNLD PKDGSVVVLG YTDRIGSDAY NQGLSERRAQ SVVDYLISKG IPADKISARG 280 290 300 310 320 | | | | | MGESNPVTGN TCDNVKQRAA LIDCLAPDRR VEIEVKGIKD VVTQPQA