Table 1 - Springer Static Content Server
advertisement

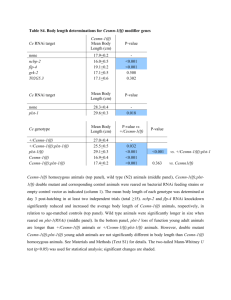
Supplementary Material to: More pronounced salt dependence and higher reactivity for platination of the hairpin r(CGCGUUGUUCGCG) compared with d(CGCGTTGTTCGCG) Margareta Hägerlöf1, Pal Papsai1, Christine S. Chow2, Sofi K. C. Elmroth1,* 10 1 Biochemistry, Chemical Center, Lund University, P.O. Box 124, SE-221 00 LUND, Sweden, and 2Wayne State University, Department of Chemistry, Detroit, MI 48202, USA Table S1 Observed first-order- and apparent second-order rate constants determined by use of HPLC technique for platination of RNAI and DNAI by complexes 1, 2, and 3 in buffered solution at pH 6.0; 5.0 mM phosphate buffer for [Na+] in the interval 19 mM ≤ I ≤ 500 mM, and 0.50 mM phosphate buffer for[Na+] = 1.0 mM. Fig. S1 A HPLC traces at different reaction times for the reaction of RNAI and 2 in 20 5 mM Na-phosphate buffer pH 6.0, [NaClO4]= 95 mM, T = 25 °C, [RNAI] = 1.25 M, CPt = 74 M. The insert shows a plot of normalized HPLC peak areas corresponding to the peak labeled as () as a function of reaction time. B HPLC traces at different reaction times for the reaction of DNAI and 3 in 5 mM Na-phosphate buffer pH 6.0, 295 mM NaClO4, T = 25 °C, 1.25 M DNAI, CPt = 74.2 M. The insert shows a plot of normalized HPLC peak areas corresponding to the peak labeled as () as a function of reaction time. Fig. S2 Observed pseudo-first-order rate constants as a function of increasing CPt for the reaction of RNAI and 2. Reaction conditions: [RNAI] = 1.25 M, 5.0 mM Na- 30 phosphate buffer, pH 6.0, 30 mM NaClO4, 50 M < CPt < 150 M (40- to 120-fold excess). Fig. S3 log(k2app) versus √I/(√I+1) as monitored for the reaction between platinum complexes 2 and 3 with RNAI and DNAI at 25 °C, pH 6.0, 19 mM ≤ [Na+] ≤ 500 mM, [Pt(II)] = 7.5 10-5 M, [oligonucleotide] = 1.25 10-6 M. Reaction of RNAI and 2 (), reaction of RNAI and 3 (), reaction of DNAI and 2 (), reaction of DNAI and 3 (). Fig. S4 A Autoradiogram illustrating rough reaction kinetics resulting from incubation of complex 3 with 3´-end labeled RNAI; CRNA = 2.2 M CPt = 120 M. The reactions were studied in Buffer A at 25 °C and were analyzed at time intervals; 0 10 min (lane 1), 1 min (lane 2), 6 min (lane 3), 20 min (lane 4), 40 min (lane 5), 1 h (lane 6), 1.5 h (lane 7), 2.5 h (lane 8), 4 h (lane 9), 6.75 h (lane 10) (20% denaturing PAGE) B Autoradiogram illustrating rough reaction kinetics resulting from incubation of complex 3 with 3´-end labeled RNAI; CRNA = 2.2 M, CPt = 220 M. The reactions were studied in Buffer A at 25 °C and were analyzed at time intervals; 0 min (lane 1), 1 min (lane 2), 6 min (lane 3), 15 min (lane 4), 36 min (lane 5), 1.1 h (lane 6), 1.5 h (lane 7), 2 h (lane 8), 3 h (lane 9), 5.5 h (lane 10) (20% denaturing PAGE) C Autoradiogram illustrating rough reaction kinetics resulting from incubation of complex 3 with 5´-end labeled DNAI; CDNA = 2.07 M CPt = 87 M. The reactions were studied in Buffer A at 37 °C and were analyzed at time intervals; 0 20 min (lane 1), 1 min (lane 2), 6 min (lane 3), 15 min (lane 4), 30 min (lane 5), 50 h (lane 6), 1h 6 min (lane 7), 1.5 h (lane 8), 2 h (lane 9), 3 h (lane 10), 4 h (lane 11) (20% denaturing PAGE). Fig. S5 A Thermodynamic data for the melt of RNAI in Buffer B (195 mM NaCl, 5 mM phosphate buffer pH 6.0, [Na+] = 200 mM), absorbance versus temperature profiles for RNA concentrations1028 M, 359 M, and 136 M B Thermodynamic data for the melt of RNAI-1 in Buffer B (195 mM NaCl, 5 mM phosphate buffer pH 6.0, [Na+] = 200 mM), absorbance versus temperature profiles for RNAI-1 concentrations 75 30 M and 9.9 M. Table S1 Observed first-order- and apparent second-order rate constants determined by use of HPLC technique for platination of RNAI and DNAI by complexes 1, 2, and 3 in buffered solution at pH 6.0; 5.0 mM phosphate buffer for [Na+] in the interval 19 mM ≤ I ≤ 500 mM, and 0.50 mM phosphate buffer for[Na+] = 1.0 mM. [Na+] / 10-3M kobs / 10-4 s-1 k2,app / M-1s-1 1 35 4.4 ± 0.4 5.9 ± 0.5 RNAI 2 1 21.8 ± 1.1 29.0 ± 1.5 RNAI 2 35 4.0 ± 0.4 5.3 ± 0.6 RNAI 2 75 2.29 ± 0.19 3.05 ± 0.25 RNAI 2 100 2.02 ± 0.21 2.69 ± 0.28 RNAI 2 300 0.96 ± 0.04 1.28 ± 0.05 RNAI 2 500 0.717 ± 0.014 0.956 ± 0.018 RNAI 3 35 59.2 ± 1.3 79.0 ± 1.7 RNAI 3 75 43.275 ± 0.5 57.7 ± 0.7 RNAI 3 100 37.8 ± 3 50.4 ± 4 RNAI 3 500 20.3 ± 2.4 27 ± 3.2 DNAI 2 1 12.1 ± 1.1 16.1 ± 1.5 DNAI 2 19 4.6 ± 0.5 6.1 ± 0.7 DNAI 2 35 3.5 ± 0.3 4.7 ± 0.4 DNAI 2 100 1.77 ± 0.13 2.36 ± 0.17 DNAI 2 300 1.6 ± 0.3 2.1 ± 0.4 DNAI 3 1 74 ± 6 99 ± 9 DNAI 3 19 30 ± 2.1 40.3 ± 2.9 DNAI 3 35 21.9 ± 0.8 29.2 ± 1.0 DNAI 3 75 10.4 ± 0.3 13.9 ± 0.4 DNAI 3 100 9.0 ± 0.6 12.0 ± 0.8 DNAI 3 300 6.9 ± 0.3 9.2 ± 0.4 DNAI 3 500 6.4 ± 0.6 8.6 ± 0.8 Target Platination oligo reagent RNAI