supplementary data - Springer Static Content Server
advertisement

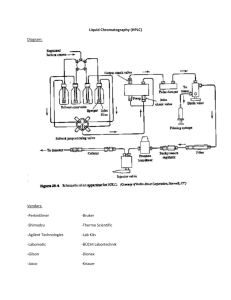
SUPPLEMENTARY DATA Conjugation of ZW800-1 with Targeted Peptides: All chemicals and solvents were of American Chemical Society or HPLC purity and were used as received. HPLC grade acetonitrile (CH3CN) and water were purchased from VWR International (West Chester, PA, USA) and American Bioanalytic (Natick, MA, USA), respectively. All other chemicals were purchased from Fisher Scientific (Pittsburgh, PA, USA) and Sigma-Aldrich (Saint Louis, MO, USA). ZW800-1 was conjugated to cyclic pentapeptide cyclo(Arg-Gly-Asp-TyrLys; cRGDyK; MW 619.7) and cyclo(Arg-Ala-Asp-Tyr-Lys; cRADyK; MW 633.7) using N-hydroxysuccinimide ester (NHS) chemistry in DMSO. Briefly, cRGDyK and cRADyK were synthesized using standard Fmoc-based solid-phase peptide chemistry and preloaded Fmoc-Gly-NovaSyn®TGT resin (Nova Biochem) in an automatic continuous flow Pioneer Peptide Synthesizer. Cleavage of protected linear peptide from the resin was performed without affecting the side chain protecting groups by using 30% of hexafluoro-2-propanol (HFIP) in dichloromethane. Head-to-tail cyclization was carried out with benzotriazole-1-yloxy-tris-pyrrolilidinophosphonium (PyBOP, Nova Biochem) (4.5 equiv), using diisopropylethylamine (9 equiv) in DMF. The crude cyclic peptide was purified by preparative HPLC. Finally, the side chain protecting groups were removed with reagent (trifluoroacetic acid/water/triisopropylsilane) (95:2.5:2.5) at room temperature for 2 h and used for further conjugation with ZW800-1 NHS ester. ZW800-1 NHS ester was prepared using highly pure ZW800-1 (> 95%) (19, 35) by adding 2 equiv of dipyrrolidino(Nsuccinimidyloxy)carbenium hexafluorophosphate (HSPyU) and 5 equiv of diisopropylethylamine (DIEA) in anhydrous DMSO. After stirring for 3 h, the reaction mixture was precipitated with an excess of anhydrous ethyl acetate. The precipitate was washed 3 times with a 50% mixture of ethyl acetate and acetone, and dried in a vacuum. 1 Purity (> 98%) was confirmed by analytical reversed-phase HPLC (770 nm absorbance) and MALDI-TOF. LC-MS Spectrometry: The purity of all compounds was measured using liquid chromatography-mass spectrometry (LC-MS) on a Waters system consisting of a 1525 binary HPLC pump with a manual 7725i Rheodyne injector, a 996 Photodiode Array (PDA) detector (Waters), and a 2475 multiwavelength fluorescence detector (Waters). The column eluate was divided in 2 using a flow splitter (Upchurch Scientific). A portion of the eluate flowed into an ELSD (Richards Scientific) while the rest flowed into a Micromass LCT ESITOF spectrometer (Waters) equipped with a Symmetry(R) C18 (4.6 x 150 mm, 5 μm) reverse-phase HPLC column. For mass spectrometry mobile phase was solvent A = 0.1% formic acid (FA) in water and solvent B = CH3CN with 95% A for 5 min and a linear gradient from 5% to 40% CH3CN (from A to B for 30 min) at a flow rate of 1 mL/min, capillary voltage was -3317V, and sample cone voltage was -50V. MALDI-TOF Mass Spectrometry: Chemical structures and purity were also analyzed using a µFocus Plate (HST, Inc., Newark, NJ) by UltraFlex III MALDI-TOF mass spectrometry (Bruker Daltonics, Inc., Billerica, MA). Desalting microcolumns, packed with Poros R2 resin (Applied Biosystems, Framingham, MA), were used to remove excess salts and contaminants from serum and urine samples. Positive and negative spectra were collected using a solid state Smartbeam™ laser operating at 100 Hz in reflectron mode (mass range 50-2,500 Da). Optical Property Measurements: All optical measurements were performed at 37˚C in PBS, pH 7.4 or 100% FBS buffered with 50 mM HEPES, pH 7.4. For measuring 2 fluorescence spectra of targeted NIR fluorophores, NIR excitation was provided by a 770 nm NIR laser diode light source (Electro Optical Components, Santa Rosa, CA) set to 10 mW and coupled through a 300 µm core diameter, NA 0.22 fiber (Fiberguide Industries, Stirling, NJ). In silico calculations of the partition coefficient (logD at pH 7.4) and surface molecular charge and hydrophobicity were calculated using MarvinSketch 5.2.1 (ChemAxon, Budapest, Hungary). Generation of a luciferase2 expressing HT-29 cell line: A plasmid containing the luciferase2 gene under the control of a CAG-promoter, a cytomegalovirus (CMV) early enhancer element and chicken β-actin hybrid promoter, pCAG-Luc2, was generated by inserting a 1.7 kb HindIII-fragment from pCAG-DEST-Frt into the pGL4.17 luciferase2 vector (Promega, Madison, WI, USA). In a parallel study, using a rat and a human colon carcinoma cell line, we have tested luciferase constructs with 4 different promoters. These studies indicated that cells transfected with the pCAG-luc2 vector showed the best luciferase expression in vivo (data not shown; manuscript in preparation). Therefore, we have used this vector to generate the HT29-luc2 cells. The human colon adenocarcinoma grade II cell line HT29 was cultured in RPMI 1640 (Invitrogen, Bleiswijk, Netherlands), supplemented with 10% Fetal Clone II (FCII; Hyclone, Thermo Scientific, Rockford, IL, USA) and 100 IU/ml penicillin, and 100 IU/ml streptomycin (Invitrogen). The HT29 cell line was transfected with the pCAG-Luc2 vector using Fugene HD (Promega). Subsequently, the cells were incubated with 500 ng/ml Genitecin (Invitrogen) and seeded single cell in a 96 well plate to establish clonal cell lines stably expressing the luciferase2 gene under the control of the CAGpromoter, HT29-luc2. To establish successful transfection, cells of 20 clones were lysed using Reporter Lysis Buffer (Promega) and luciferase expression was measured using the Luciferase assay system (Promega) and a Wallac 420 microplate reader (Perkin Elmer). 3 Four clones with the highest luciferase expression were tested for tumorigenic capacity in vivo by subcutaneous injection of 5 x 105 cells at the back of nude mice. Tumor growth was monitored weekly using an IVIS Spectrum imaging system (Caliper LifeSciences, Hopkinton, MA, USA). Results showed that clone 1 induced the strongest tumor growth and was used for further experiments (data not shown). Intraoperative Near-Infrared Imaging System (FLARETM): The Fluorescence-Assisted Resection and Exploration (FLARETM) NIR imaging system used in this study has been described in detail previously in the Annals of Surgical Oncology by Troyan et al. Briefly, the system was positioned using an articulated arm anywhere over the surgical field, with a working distance of 45 cm above the subject. White light (400–650 nm) and NIR fluorescence excitation light (745–779 nm) were generated by light-emitting diodes over a 15-cm diameter projection. Color video (surgical anatomy) and NIR fluorescence (fluorophore distribution) images were acquired simultaneously via custom-designed optics, and displayed individually or merged. For merged images, the grey-scale NIR fluorescence image was pseudocolored in lime green and superimposed on the color video image. Fluorescence intensity of any desired region of interest (ROI) was quantified using a 12-bit pixel scale (range 0–4095). 4