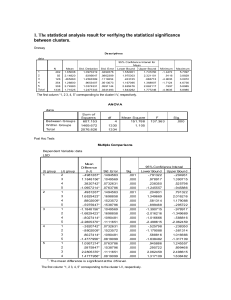
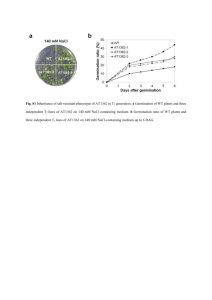
gene confers resistance to imidazolinone herbicides in China
advertisement

1 Supplementary information 2 Fig. S1 Subcellular localization of the BnAHAS1 protein. The SE5-GFP fusion protein or GFP alone 3 expressed under the control of the CaMV 35S promoter in Arabidopsis protoplasts was observed using 4 confocal microscopy. The photographs were taken in the blue channel (left), the red channel (middle), and 5 merged (right). Scale bars represent 10 µm. 6 7 9 35S::GFP 8 GFP 11 12 13 14 15 16 17 18 19 20 21 22 35S::BnAHAS1-GFP 10 Chlorophyll Merged 23 Fig. S2 Diagram of the procedure of F1 seeds using the restore line 10M169 with IMI resistance to enhance 24 hybrid seed purity in hybrid rapeseed production. The allelic composition of IMI resistant for the restore line 25 10M169 and the CMS line Ning A7 were represented by RR and rr in round brackets, respectively, and the 26 resulting F1 hybrid was represented by Rr; the off-type contaminants such as sibs/selfs produced by trace 27 pollen and biological mixtures were also represented by rr. 28 29 30 The CMS line Ning A7 (rr) × The restore line 10M169 (RR) 31 32 33 Artificial hybridization Hybridization in tents Natural pollination 34 35 36 F1 (Rr) biological mixtures (rr) F1 (Rr) 39 40 41 42 43 44 45 F1 (Rr), sibs/selfs (rr), and After IMI treatment 37 38 F1 (Rr) and sibs/selfs (rr) 0 F1 (Rr) F1 (Rr) 46 Table S1 List of primers used for amplification and genotyping of the AHAS genes 47 Primer or probe Sequence (5'-3') Use AHAS1-F1 TCAAGAACAGTTAGATCCAC Gene cloning AHAS1-R1 GATCACCAGCTTCATCTCT Gene cloning AHAS2-F1 AAGCAATTTCTCGCAACACTC Gene cloning AHAS2-R1 CAGAAGAGAGCATAGAATAATCAA Gene cloning AHAS3-F1 CTCTCTCTCTCTCATCTAACCAT Gene cloning AHAS3-R1 ACTGAAACTAAGTCTTTT ACCAT Gene cloning KpnI-AHAS1R-F* CGCGGTACCTCATCTCTCTCTCCTCTAACC Plasmid construction EcoRI-AHAS1R-R* GTCCGGAATTCTCAGTACTTAGTGCGACCATCCCCTTC Plasmid construction AHAS1-F2 TTCTCCTTAACCCCACAGAAAGA RT-PCR AHAS1-R2 GGGAGCGTAGCGGGAGAC RT-PCR AHAS1-P Fam+CCGTCAATGTCGCACCTC CTTCC+Tamra RT-PCR AHAS2-F2 CTTCGTTTTCGTTCTTCGGC RT-PCR AHAS2-R2 AGACACGAGT AGCACGGCG RT-PCR AHAS2-P Fam+CAAAAGCTTCCGTCTTCTCCCTGCC RT-PCR AHAS3-F2 CTCCTTAACCCCACAGAAACC RT-PCR AHAS3-R2 GGGAGCGTAGCGGGAGAT RT-PCR AHAS3-P CAACTCACCCGTCAATGTCGCACC RT-PCR 18s rRNA-F AACGG CTACCACATCCA RT-PCR 18s rRNA-R CACCAGACTTGCCCTCCA RT-PCR 18s rRNA-P Fam+AGCAGG CGCGCAAATTACC RT-PCR NcoI-AHAS1-F* AGCTCCATGGCGGCGGCAACATCGTC Plasmid construction SpeI-AHAS1-R* CAAGACTAGTGGAGATGGCGAGTGGACGGTG Plasmid construction AHAS1-S** CATCTTTGAAAGTGCCACAAC AP-PCR AHAS1-R** CATCTTTGAAAGTGCCACAAT AP-PCR AHAS1-C CTTTCGCTAGCAGGGCTAAA AP-PCR 48 49 * The underlining represents the enzyme sites. 50 ** The underlining shows mismatched bases. The bold bases indicate SNP loci. 51 52 53 54 Table S2 Genotype frequencies at SNP (G/A) sites in the F2, BC1, and BC2 populations 55 56 Genotype Cross χ2-value P-value 39 154 0.20 0.50–0.75 117 0 227 0.45 0.25–0.50 98 113 201 0.37 0.50–0.75 G/A A/A F2 (3075R×M9) 35 80 BC1 [(3075R×M9) ×3075R] 110 0 BC2 [(3075R×M9) ×M9] 57 No. of plants G/G