file - SpringerPlus
advertisement
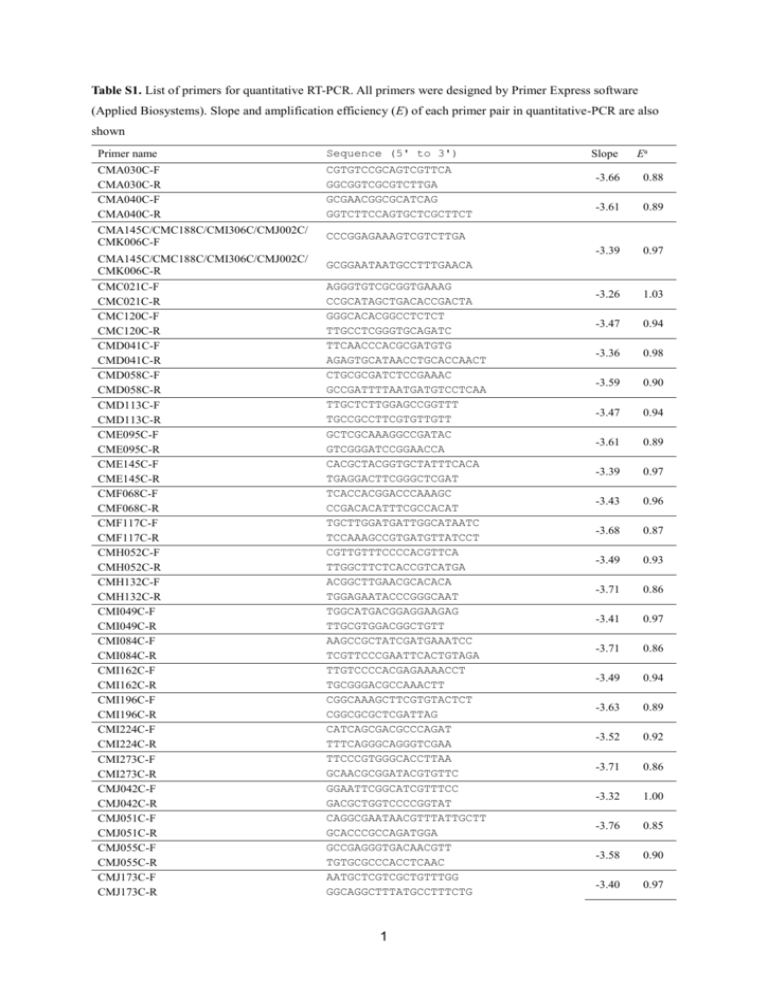
Table S1. List of primers for quantitative RT-PCR. All primers were designed by Primer Express software (Applied Biosystems). Slope and amplification efficiency (E) of each primer pair in quantitative-PCR are also shown Primer name Sequence (5' to 3') CMA030C-F CMA030C-R CMA040C-F CMA040C-R CMA145C/CMC188C/CMI306C/CMJ002C/ CMK006C-F CGTGTCCGCAGTCGTTCA GGCGGTCGCGTCTTGA GCGAACGGCGCATCAG GGTCTTCCAGTGCTCGCTTCT CMA145C/CMC188C/CMI306C/CMJ002C/ CMK006C-R CMC021C-F CMC021C-R CMC120C-F CMC120C-R CMD041C-F CMD041C-R CMD058C-F CMD058C-R CMD113C-F CMD113C-R CME095C-F CME095C-R CME145C-F CME145C-R CMF068C-F CMF068C-R CMF117C-F CMF117C-R CMH052C-F CMH052C-R CMH132C-F CMH132C-R CMI049C-F CMI049C-R CMI084C-F CMI084C-R CMI162C-F CMI162C-R CMI196C-F CMI196C-R CMI224C-F CMI224C-R CMI273C-F CMI273C-R CMJ042C-F CMJ042C-R CMJ051C-F CMJ051C-R CMJ055C-F CMJ055C-R CMJ173C-F CMJ173C-R GCGGAATAATGCCTTTGAACA Slope Ea -3.66 0.88 -3.61 0.89 -3.39 0.97 -3.26 1.03 -3.47 0.94 -3.36 0.98 -3.59 0.90 -3.47 0.94 -3.61 0.89 -3.39 0.97 -3.43 0.96 -3.68 0.87 -3.49 0.93 -3.71 0.86 -3.41 0.97 -3.71 0.86 -3.49 0.94 -3.63 0.89 -3.52 0.92 -3.71 0.86 -3.32 1.00 -3.76 0.85 -3.58 0.90 -3.40 0.97 CCCGGAGAAAGTCGTCTTGA AGGGTGTCGCGGTGAAAG CCGCATAGCTGACACCGACTA GGGCACACGGCCTCTCT TTGCCTCGGGTGCAGATC TTCAACCCACGCGATGTG AGAGTGCATAACCTGCACCAACT CTGCGCGATCTCCGAAAC GCCGATTTTAATGATGTCCTCAA TTGCTCTTGGAGCCGGTTT TGCCGCCTTCGTGTTGTT GCTCGCAAAGGCCGATAC GTCGGGATCCGGAACCA CACGCTACGGTGCTATTTCACA TGAGGACTTCGGGCTCGAT TCACCACGGACCCAAAGC CCGACACATTTCGCCACAT TGCTTGGATGATTGGCATAATC TCCAAAGCCGTGATGTTATCCT CGTTGTTTCCCCACGTTCA TTGGCTTCTCACCGTCATGA ACGGCTTGAACGCACACA TGGAGAATACCCGGGCAAT TGGCATGACGGAGGAAGAG TTGCGTGGACGGCTGTT AAGCCGCTATCGATGAAATCC TCGTTCCCGAATTCACTGTAGA TTGTCCCCACGAGAAAACCT TGCGGGACGCCAAACTT CGGCAAAGCTTCGTGTACTCT CGGCGCGCTCGATTAG CATCAGCGACGCCCAGAT TTTCAGGGCAGGGTCGAA TTCCCGTGGGCACCTTAA GCAACGCGGATACGTGTTC GGAATTCGGCATCGTTTCC GACGCTGGTCCCCGGTAT CAGGCGAATAACGTTTATTGCTT GCACCCGCCAGATGGA GCCGAGGGTGACAACGTT TGTGCGCCCACCTCAAC AATGCTCGTCGCTGTTTGG GGCAGGCTTTATGCCTTTCTG 1 CMJ250C-F CMJ250C-R CMJ272C-F CMJ272C-R CMJ293C-F CMJ293C-R CMJ305C-F CMJ305C-R CMK041C-F CMK041C-R CMK131C-F CMK131C-R CMK188C-F CMK188C-R CML036C/CML059C/CMM231C-F CML036C/CML059C/CMM231C-R CML209C-F CML209C-R CMM068C-F CMM068C-R CMM167C-F CMM167C-R CMM196C-F CMM196C-R CMM299C-F CMM299C-R CMN017C-F CMN017C-R CMN233C-F CMN233C-R CMN285C-F CMN285C-R CMO121C-F CMO121C-R CMO124C-F CMO124C-R CMO128C-F CMO128C-R CMO229C-F CMO229C-R CMO245C-F CMO245C-R CMO276C-F CMO276C-R CMO291C-F CMO291C-R CMP129C-F CMP129C-R CMP193C-F CMP193C-R CMP260C-F CMP260C-R CMQ172C-F CMQ172C-R CMQ191C-F CMQ191C-R CTACCCGAAGCTTGACGGTAA GATACAAGGGCCACCGGTATC CGTTGTCCAGGTTCCAACGT CGTGTGCGCTTTCGTATGC GCGCGCTCTCGACATTCT CACGCTGTCGAGCAGTTCAG CGGACAAGGTATCGCATATCTCA CCCGCCCCTCGATTAACT CGGCAAGTCGGCGTCTT CGACGCCAAAACGAATGTC CGATCTCCTGGTTACGAATATCG CGTTGCAGGCGCGTTT TGTCAAGAGCTCGGCTACGTACT GAGCATGCATTCCGCGTTA GCAGGTCGCGCAATATCG GTGCAAAGTTCTGACCCATGAC ACGATGGCCAGATGGATGA TAGCGCCGCCGTCACT CATCAGCGGTGTCGGAATG TCCTGAGGTACGCGTGCAT TCGAGTCCACTGGCGTGTT CGCACCGCCTTTCAGATG TGCCGGCAAGTCAAATACC GCCCACGGAACGAAACTTT TGTAGCCCTGCCCATGCT AGCGCTCCTGTCGACGAT CACTTTCGGAGGTGCTTTGC CCGGCATACGGAGCTTGAT TCATTGCGTCGCCGTATG GCAGGGTACGCAGGTCAATT CTCGTATGCGATGCTTTTGG GCAGCCTGGAGTGGTGTCA TGCGCGGAGCGTACGT GGATCACATCCGGGTGCTT TGCCGATGGTGGAGAAGTTC CCCGACCCAGTCCCAAA TCAAGGATTGTGCGGCAAA GCATTGCGCCGGAGACT CCGTCGTTGCTCTCTTGTGA GTCGAGCACGCGTTTCG CCCGAGGACGCGGATATAA TCGCACCCGGTTGCA CCCCGACGCTGGTATGAG AAACAATCCGCTCGGTATGC CCATAGACGGAGCGGATGAG GCACCACCGCGTCCTTT GAAGCACAGGGCGACTTGAC TCGATGACCTTACAGGCAAGCT GTGCGTTTCCGCTGCTAAA GCATCCGCAAAGCCCTTAT CGACGGCACCGATTGC TGCACCGGATAGAGACCTTTG GCCATCGGTTTATGCCATGT TGCAGTGCTACCCCGAAATC GGGAACGATTCGGTGAAGAA ATCCCACCGTAGGCCTGATT 2 -3.38 0.98 -3.57 0.90 -3.63 0.89 -3.54 0.92 -3.56 0.91 -3.51 0.93 -3.29 1.01 -3.46 0.95 -3.55 0.91 -3.51 0.93 -3.31 1.00 -3.65 0.88 -3.60 0.89 -3.71 0.86 -3.53 0.92 -3.43 0.96 -3.67 0.87 -3.59 0.90 -3.61 0.89 -3.75 0.85 -3.58 0.90 -3.76 0.85 -3.47 0.94 -3.60 0.90 -3.64 0.88 -3.83 0.82 -3.49 0.93 -3.72 0.86 CMQ234C-F CMQ234C-R CMR014C-F CMR014C-R CMR476C-F CMR476C-R CMS195C-F CMS195C-R CMS272C-F CMS272C-R CMS327C-F CMS327C-R CMT034C-F CMT034C-R CMT209C-F CMT209C-R CMT216C-F CMT216C-R CMT256C-F CMT256C-R CMT285C-F CMT285C-R CMT362C-F CMT362C-R CMT412C-F CMT412C-R CMT497C-F CMT497C-R CMT561C-F CMT561C-R CMT582C-F CMT582C-R CMT611C-F CMT611C-R CMT633C-F CMT633C-R CMV013C-F CMV013C-R CMV014C-F CMV014C-R CMV153C-F CMV153C-R CMV154C-F CMV154C-R CMW001C-F CMW001C-R CMW002C-F CMW002C-R a GGCTGCGTTCCGTCAAAA TTCGAAGCTCCCGGACTTT CCGCGACATCATGCAGAAT CCGGCTGTTCCATCGTAAAG GGCTGCTGGTTATGTGTTCATG CGCTCTCCACGAGAAAAGCT TCGCCGCATGGAATGAA TTGAGGATGTTGGCCGTTATC ATTCCGAGCAATGTTCTGGATT CAGCGGCCCCTTGATG AGAAGACGCCAAGGGACTTCT TTCAAGCACACAAACGGGATT GGCAGCAACCAAGGAGGAA TCGCGCTGCGCTTGA TTGTGGATGCAGCAAAACGT GCCTTCGAGCCGAACAATC CGGACGACGGCGTTACC TTAACCGCTGCGTGATGCT AGGCCCGAGCGGAGAA CAGTGCGAACGGTGAAAGTTC GCGATTATGCCGTGCTTGA CGTAAAATCATCGGCAACGA GCACCGGATGTCTTCCAAA CCGGAACACTCGGATTACAGA TTGTTGATGCGACCTGTATGC GGTTCGGCAACAGTACAACGT CGGCCTTAACCCGGAAA CGCGGTCGTGAAGGTTTT GCATGCGGGCCCTGTA TGTTTTTGACGCCCTTTGGT CGTCCTTCCGACGGTTCA GGACTTCACCTCGCCAGTTG GGCCATCGGCGACCAT CGATCTCGTTTTCCGTTTGG CGCATGGGTGTGGAAAGG ATGGAAGGTGCAACCACAACT GCATCTGGCGAAGTCAAAGG CGCAAATTCTGCTCTCTTATAACATT TGGCATTCAACTCGACTAAAGG TTCGTTTGCAGGTCGATTCA ATGGCCGAATTATTCGGAAA GGTGCTGAAAATAAATGCATCGA TGCAGGCATGTTGCATTATACA TCCGGGACCTCGAATCACTA TGTTTTTCTTTAAATTTTATTCGCCTACT CTTTATTAAATAACCATAAAAAACTTACACACATT TTGGTGGAATGGTGATCGTTATT GCGCTGTCACGACTATCTATAATCC -3.66 0.88 -3.57 0.91 -3.45 0.95 -3.62 0.89 -3.39 0.97 -3.78 0.84 -3.67 0.87 -3.62 0.89 -3.46 0.94 -3.33 1.00 -3.62 0.89 -3.54 0.92 -3.44 0.95 -3.55 0.91 -3.54 0.92 -3.60 0.90 -3.39 0.97 -3.69 0.87 -3.85 0.82 -3.44 0.95 -3.83 0.82 -3.24 1.04 -3.46 0.94 -3.58 0.90 Amplification efficiency (E) for each gene was determined from following equation: E = 10-1/Slope -1. 3 Figure S1 Changes in cell size and shape of C. merolae cell in the flat-plate culture. (a) A plot for areas of a plastid and an extraplastid at 0, 1, 3, and 6 h in the culture. Compartmentation of the plastid and extraplastid is explained in Figure 1f. Plots for lateral and longitudinal diameters of a plastid (b) and an extraplastid (c). Areas and diameters were measured for 30 cells in each culture time 4 Figure S2 Changes in expression level of genes involved in central carbon metabolism in the flat-plate culture. The transcript level of each gene was measured by quantitative RT-PCR, and corrected using transcript levels of 18S rRNA gene as an internal standard, and then normalized to the transcript level of each gene at 0 h. Genes are classified into six classes with respect to the peak of transcript level; genes of <2-fold change (a), 2 to 4-fold changes (b), 4 to 8-fold changes (c), 8 to 16-fold changes (d), 16 to 32-fold changes (e), and >32-fold change (f). Each value is an average ± standard deviation of three independent assays 5 Figure S3 Culture under the darkness with addition of organic substances. C. merolae was cultured in a flask with rotary-shaking under the dark condition with addition of 200 mM glucose, 200 mM glycerol, 200 mM succinic acid, or 5 mM L-lactic acid for four weeks and OD750 was measured every one week. In the assay, the cells were exposed to room light at the sampling time for a short period of time. 6