Inversion loops will form in inversion heterozygotes
advertisement

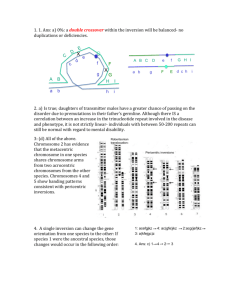
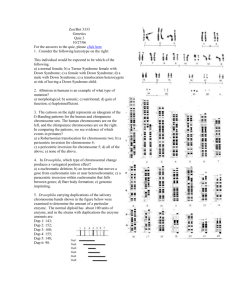
Inversion loops will form in inversion heterozygotes. A cross over within the inversion loop will produce duplications and deficiencies in the resultant recombinants. These would give rise to cells that might not complete gametogenesis or would produce aneuploid fertilization products. 1. Numbers 2 and 3 are inversion heterozygotes. Ans (b). 2. If the two homologs are both inversions, then the loci should pair up along their length and no loops would form during 1st meiotic prophase. Crossovers that occur between homologs 1 and 4 would not generate duplication and deletion gametes because no inversion loop would form. The other two nonsister chromatids would segregate without crossovers. Ans (d) 3. Both 3 and 4 are inversion heterozygotes, but only 3 contains a paracentric inversion. Therefore, only 3 would generate acentric and dicentric fragments if a crossover occurred in the inversion loop. Ans: none of the above (e) 4. Nondisjunction in the homozygous parent (father) can give rise to an aa gamete. What about heterozygote? Suppose cell undergoes nondisjunction in 2nd meiotic metaphase: a a Both heterozygote (mom) and dad can undergo nondisjunction to yield a trisomic child…Ans: (c) 5. b+ a- Deletion ‘uncovers’ recessive mutation: will see an a- mutant phenotype in deletion heterozygote (pseudodominance; p. 496) b- If area of deletion does not contain b gene, then a wild type copy of b will be on deletion chromosome…phenotype of heterozygote will be wild type Deletion heterozygotes can therefore be used for mapping…. deletion 1: deletion 2: deletion 3: deletion 4: a,d,e c,d,f b,c d,e Start with deletions covering smallest area b must be next to c d must be next to e (bcdf) or (bcfd) (eda) or (dea) or (ade) bc and de bcfdea or eadfcd the consistent possibility Ans: (c) 6. As discussed in class, females have a greater chance of passing on the disorder than males- they are more likely to amplify a premutation to levels that lead to symptoms. The other statements are not true, because premutation alleles are subject to variable expressivity. Please see pp. 216-17 in text. Ans: (a) 7. False. F+ x F+ or Hfr x Hfr or F+ x Hfr will not yield recombinants. Only donor x recipient crosses will. 8. The key to this question is to remember that F– cells can produce either zero (0), low numbers (L) or many recombinants (M), depending on what strain they are crossed to: So strains 3, 6, and 8 must be F– strains. Given this information, strains 2 and 7 produce many recombinants. They are the Hfr strains. Strains 1, 5 and 4 are F+ strains. Ans: (d) 9. Agar Type 1 2 3 4 Str A B C D + + + + + + + + + + + + + + + + - Timings of Samples 0 2.5 5 7.5 10 12.5 15 17.5 20 25 Number of Colonies on Agar of Type: 1 B 2 A 3 C 4 D 0 0 0 0 0 0 0 0 15 0 0 0 104 0 0 0 215 0 8 0 301 6 75 0 347 67 150 5 400 104 202 50 402 150 205 90 398 153 203 90 Note: Plate 1 selects for the transfer of the b+ gene, since it does not contain nutrient B, Plate 2 selects for the transfer of the a+ gene, since it does not contain nutrient A, etc. 9. The time of entry map indicates the order is bcad. The b+ gene is transferred first, since b+ exconjugants appear earliest(~ 5 min), and in the greatest overall frequency. This is followed by c+ (~10 min), then a+ (~12.5 min) then d+ (~15 min). Ans: none of the above (e) 10. False, the distance between b and c is about 5 min. The distance between c and a is about 2.5 min.