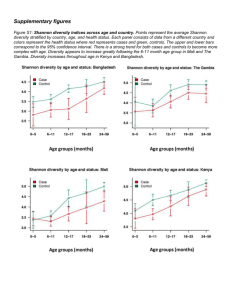
significant abundant
advertisement

Supplementary Materials for “Coral and macroalgal exudates vary in neutral sugar composition and differentially enrich bacterioplankton populations in a tropical reef ecosystem” by Craig E. Nelson, Stuart J. Goldberg, Linda Wegley Kelly, Andreas F. Haas, Jennifer E. Smith, Forest Rohwer, and Craig A. Carlson Supplementary Methods: Compositional analysis of dissolved combined neutral sugars – The concentrations of DCNS were determined using established methods (Goldberg et al. 2009, 2010). Surface reference waters (SRW; collected in the Santa Barbara Channel) and Nanopure® blanks were analyzed in triplicate, and unknown samples were analyzed in duplicate (see Goldberg et al. 2009 for details about the use of SRW). Four mL aliquots of Nanopure® water blanks and unknown samples were transferred into individual 5 mL pre-combusted (450 °C for ~4 hrs) glass ampoules (Wheaton). All samples were acidified with H2SO4 (Cf=0.85 M) and hydrolyzed for 24 h at 100 °C. At the completion of hydrolysis, the samples were cooled to room temperature and neutralized with CaCO3 in clean polycarbonate (PC) tubes. These PC tubes were vortexed vigorously prior to a 30-minute centrifugation at 28,760 x g. The sample supernatants were collected and then transferred into combusted glass 20 mL scintillation vials. Neutralized samples were desalted using an equal mixture of anion (AG 2X-8, 20-50 mesh; Bio-Rad) and cation (AG 50 WX-8, 100-200 mesh; Bio-Rad) exchange resin. The DCNS were analyzed by high performance liquid chromatography and pulsed amperometric detection (HPLC-PAD). Chromatographic conditions for the separation of monosaccharides followed the procedures of Goldberg et al., (2009) after slight modification. After the time period of peak integration (20 minutes at 12 mM NaOH), there were 10 min and 30 min washes with 200 mM 12 mM NaOH, respectively. Fucose, rhamnose, arabinose, galactose, glucose, and mannose + xylose (co-elute due to column chemical characteristics) were separated using a CarboPac PA-10 analytical and guard columns (Dionex; Sunnyvale, CA). Within any given sequence of samples, a known monostandard mix of 6 monosaccharides (Dionex; Sunnyvale, CA) was analyzed after every 6th sample to monitor the performance variability of the working electrode, reference electrode, and the CarboPac columns. A quantification standard consisting of a mix of the 7 target monosaccharides was used, and was also hydrolyzed and desalted. High (3000, 2000, 1000, and 600 nM) and low (500, 250, 100, 50, and 20 nM) concentration standard curves were analyzed 2 to 3 times during each sequence. The recovery of individual monosaccarides after desalting and acid hydrolysis was 70-90% and ~55-60%, respectively. The coefficient of variability for the concentrations of DCNS and individual monosaccharides within SRW were comparable to historical values (see Goldberg et al., 2009). The coefficients of variability for the concentrations of DCNS within SRW ranged between 3 and 5% while those for fucose, rhamnose, arabinose, galactose, glucose, and mannose + xylose were ~6%, 15%, 16%, 3%, 5%, and 4%, respectively. DNA extraction, amplification, and pyrosequencing - Frozen Sterivex (n=3 replicates from each treatment, n=4 controls, and n=6 ambient samples) were thawed 30 minutes at 37°C and lysis proceeded by 12 h 55°C digestion with sodium dodecyl sulfate (1%) and Proteinase K (0.2 g L-1) and genomic DNA extraction from 15% of the total lysate using silica microcentrifuge columns (Qiagen DNEasy – Tissue protocol). The polymerase chain reaction (PCR) was used to construct multiplex amplicon pyrosequencing libraries of the 16S ribosomal subunit gene using primers 8f and 338r as described previously (Nelson and Carlson 2012). PCR reaction conditions were 270s 94°C hotstart, 35 cycles of 30s 94°C, 45s 57°C, 90s 72°C, 600s 72°C extension and 4°C storage. PCR products were pooled at equimolar quantities and pyrosequenced on a Roche/454 GS FLX using Titanium Chemistry (laboratory of Stefan Schuster, Pennsylvania State University). Phylogenetic community analysis – Amplicons of community 16S rRNA gene sequences were dereplicated, aligned, clustered into operational taxonomic units (OTUs), and analyzed for diversity and phylogenetic similarity among samples as within the software environment MOTHUR v20 (Schloss et al. 2009) as previously described (Nelson and Carlson 2012). Median read lengths were ~350bp and most reads spanned the full amplicon. An average of 607 quality sequences were acquired from each sample (range 154-1439). A distance matrix was built and sequences were average-neighbor hierarchically clustered into 95% sequence identity OTUs (95% similary in the V1-V2 amplicon region is estimated to roughly approximate 97% similarity over the entire 16S subunit gene; Schloss 2010). Each OTU was consensus-classified (70% minimum confidence) and a consensus sequence for each OTU was built to represent 95% of all sequences in the OTU cluster. Rare OTUs with less than 5 sequences among the 22 samples were removed from further analysis. Weighted Unifrac analysis (Lozupone and Knight 2005) was used to calculate community similarity among samples. A maximum-likelihood phylogenetic tree was constructed from 180 consensus sequences (one representative of each OTU) using a gamma-distributed generalized time-reversible nucleotide evolution model with 100X rapid-bootstrapping (RAxML; Stamatakis 2006). These final weighted Unifrac distance matrices were verified to have strong correlation (rMantel > 0.85, p < 0.01) with distance matrices derived from alternate analytical frameworks: 1) Unifrac matrices generated either unweighted (147,230 sequences), weighted by unique sequences (31,539 sequences) or sequences clustered at the 95% OTU level (3,138 sequences) or from 2) Bray-Curtis distances among samples calculated from relative abundances of either OTUs or clade- and family-level consensus-classified phylotypes. Moreover, these correlations persisted under either SINA or MOTHUR alignment algorithms and using either relaxed neighbor-joining or maximum likelihood tree-building programs (Clearcut or RaxML), emphasizing that dominant patterns in community differentiation persisted regardless of community distance metric (Unifrac vs. Bray-Curtis), phylogenetic resolution (sequence aggregation at unique, 95%, or classification-based phylotyping levels), alignment algorithm (SINA or MOTHUR), or phylogeny algorithm (Clearcut or RAxML). In order to more accurately classify sequences according to phylogenetic position (rather than assigning phylotypes solely based on the consensus SILVA Bayesian taxonomic classification described above) we built a maximum likelihood tree (as above) scaffolded with the nearest full-length 16S SINA-aligned neighbors (172) to each OTU consensus sequence (180) downloaded from the SILVA v106 SSU Ref database. Using the consensus taxonomy as a guide, clades were identified within the tree according to high bootstrap confidence areas (>0.7) and visual assessment of tree structure, branch length, and consistency of Bayesian consensus classification described above. Clades were named according to the lowest common identifier in consensus classifications of OTUs and scaffold reference sequences. All clade-level phylotype analyses were conducted by summing relative abundances of common OTUs grouped manually according to clades in this scaffolded maximum-likelihood phylogeny. Supplementary Results: Bacterial populations selected against in exudate treatments - Of the 27 OTUs with significant ANOVA’s indicating differences in relative abundance among treatments, a suite of 6 OTUs belonging to families commonly abundant in oligotrophic oceanic waters showed steady and significant declines from the ambient to the control to the various exudate treatments, including Chloroplasts (OTU85, removed from analysis), Synechococcus (OTU22) and four abundant OTUs of common marine oligotrophs belonging to the clades SAR11 (OTU9 and OTU43), SAR116 (OTU15), and the NS9 clade of Flavobacteria (OTU38; Table 5). Notably, two OTUs which were among the most common in ambient waters remained relatively abundant in all of the dilution cultures, one belonging to the Alteromonadaceae NOR5 clade (OTU5; 1.95% in ambient and 1.17 to 3.72% in cultures) and another belonging to the Flavobacterial NS5 clade (OTU7; 2.15% in ambient and 3.47-8.22% in cultures, with the exception of the Porites culture – 0.10%). All of the incubation cultures, including the controls, were dominated (20.2% to 33.5%) by a single OTU belonging to the Alphaproteobacterial family Rhodobacteraceae (OTU1, 96.7% identity to Phaeobacter) which was also common in ambient waters but at significantly lower relative abundance (8.0%). In addition, there were seven less abundant taxa that were significantly enriched in multiple incubation bottles (including the control) relative to the ambient waters (Supplementary Table S7; none of these taxa showed statistically significant differences among treatments). Diversity patterns in bacterioplankton communities – Diversity indices were calculated from OTU abundance data (Supplementary Table S8) using MOTHUR to calculate Shannon diversity index, Simpson evenness index, Chao1 nonparametric richness estimates, and to implement the mixed-model parametric richness estimation routine CatchAll (Bunge 2011; McCliment et al. 2011). Treatments did not differ significantly in mean sequence reads acquired per sample (ANOVA p = 0.17, Supplementary Figure S9). However, although reads per sample did not covary with Shannon or Simpson diversity indices (p > 0.05), both observed and estimated richness of OTUs covaried strongly with reads per sample (p < 0.001); we therefore included reads per sample as a random-effect variable in ANOVA models comparing Richness among treatments. The number of OTUs observed differed significantly among treatments (p = 0.011), with highest counts in the Porites exudate treatment and ambient water and significantly lower counts in the Amansia treatment (Tukey α = 0.05). Richness was also significantly different among treatments using either nonparametric (Chao1) or parametric (CatchAll) estimators (ANOVA p = 0.033 and 0.038, respectively). The Chao1 estimator only found a significantly higher richness in the Ambient water (p = 0.011) while the CatchAll estimator only found a significantly higher richness in the Porites amendment treatment (p = 0.015), and all other treatments did not differ significantly (p > 0.05). Shannon and Simpsons indices of diversity differed strongly among treatments (p < 0.001), with both showing lower diversity and evenness in the Amansia treatment relative to all other treatments (Tukey α = 0.05). We found no significant differences in diversity metrics among control, Turbinaria, and Halimeda treatments (Supplementary Figure S9). Supplementary References: Bunge, J. 2011. Estimating the number of species with catchall. Pac Symp Biocomput 121-130 , doi:10.1142/9789814335058_0014 Goldberg, S. J., C. A. Carlson, B. Bock, N. B. Nelson, and D. A. Siegel. 2010. Meridional variability in dissolved organic matter stocks and diagenetic state within the euphotic and mesopelagic zone of the North Atlantic subtropical gyre. Marine Chemistry 119: 9-21 , doi:10.1016/j.marchem.2009.12.002 Goldberg, S. J., C. A. Carlson, D. A. Hansell, N. B. Nelson, and D. A. Siegel. 2009. Temporal dynamics of dissolved combined neutral sugars and the quality of dissolved organic matter in the Northwestern Sargasso Sea. Deep Sea Research Part I: Oceanographic Research Papers 56: 672-685 , doi:10.1016/j.dsr.2008.12.013 Lozupone, C., and R. Knight. 2005. UniFrac: a New Phylogenetic Method for Comparing Microbial Communities. Appl. Environ. Microbiol. 71: 8228-8235 , doi:10.1128/AEM.71.12.8228-8235.2005 McCliment, E. A., C. E. Nelson, C. A. Carlson, A. L. Alldredge, J. Witting, and L. A. AmaralZettler. 2011. An all-taxon microbial inventory of the Moorea coral reef ecosystem. ISME J. Nelson, C. E., and C. A. Carlson. 2012. Tracking differential incorporation of dissolved organic carbon types among diverse lineages of Sargasso Sea bacterioplankton. Environmental Microbiology 14: 1500-1516 , doi:10.1111/j.1462-2920.2012.02738.x Schloss, P. D. 2010. The Effects of Alignment Quality, Distance Calculation Method, Sequence Filtering, and Region on the Analysis of 16S rRNA Gene-Based Studies J.A. Eisen [ed.]. PLoS Computational Biology 6: e1000844 , doi:10.1371/journal.pcbi.1000844 Schloss, P. D., S. L. Westcott, T. Ryabin, J. R. Hall, M. Hartmann, E. B. Hollister, R. A. Lesniewski, B. B. Oakley, D. H. Parks, C. J. Robinson, J. W. Sahl, B. Stres, G. G. Thallinger, D. J. Van Horn, and C. F. Weber. 2009. Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Applied and Environmental Microbiology 75: 7537-7541 , doi:10.1128/AEM.01541-09 Stamatakis, A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688 -2690 , doi:10.1093/bioinformatics/btl446 Supplementary Figure and Table Legends: Supplementary Figure S1. Example photographs of the four benthic primary producers found in lagoonal reefs on the north shore of Moorea, French Polynesia used to produce exudates in this study. Species are as follows: a)Turbinaria ornata – Ochrophyta; b) Halimeda opuntia – Chlorophyta; c) Amansia rhodantha – Rhodophyta; d) Porites lobata - Cnidaria. Photos a-c sourced from AlgaeBase: Guiry, M.D. & Guiry, G.M. 2011. http://www.algaebase.org, National University of Ireland, Galway; photo d sourced from MCRLTER at http://mcr.lternet.edu/education with respective copyrights: a) Eric Coppejans(eric.coppejans@ugent.be); b) John Huisman (j.huisman@murdoch.edu.au); c) Heroen Verbruggen (heroen.verbruggen@gmail.com); d) Matthew Meier (matt@matthewmeierphoto.com). Supplementary Figure S2. Replicate bacterioplankton growth curves over time in each treatment. Supplementary Figures S3. Complete phylogeney of all OTUs analyzed in this study and nearest neighbors. Bars at right of each OTU give relative abundance of each taxa in each treatment. Supplementary Table S4. Fifteen most common OTUs in each treatment, with mean relative abundances. A set of six OTUs which were common to most treatments are colorcoded for ease of visualization of shifts in dominance from Ambient Waters to experimental treatments. Supplementary Table S5. OTU mean relative abundances among treatments with ANOVA and Dunnet’s tests for significant differences among treatments. ANOVA p-values in bold italic are significant after multiple comparison corrections. Mean relative abundances significantly different from Control treatments (Dunnet’s p < 0.05) are underlined; those in bold are higher while those in italics are lower than Control cultures. *Note that for OTUs statistical tests did not include Ambient comparisons. Supplementary Figure S6. Maximum-likelihood phylogeny of selected OTUs differing significantly among treatments and their nearest cultured isolate neighbors. Supplementary Table S7. Eight taxa which were enriched in all dilution culture incubations (exudate-amendments and controls) relative to ambient waters. Supplementary Table S8. Sequence reads, observed OTUs, diversity indices, and richness estimates for each sample. Multiplex barcodes are listed to associate each sequence set with the SRA Accession. *Simpson Diversity here is calculated as 1-D. Supplementary Figure S9: Mean OTU diversity indices among treatments. Whiskers represent one standard error of the mean. Data are listed in Table S8.