Comparative Mapping - Cattle Genome Database
advertisement

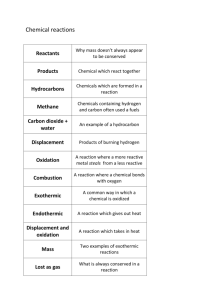
Table 4. This table is cited in the paper Barendse, W. et al. Mammalian Genome 8, 21 - 28. The locations of the genes mapped by linkage analysis in cattle, with comparisons to humans, mice, rats and pigs. The number of conserved syntenies and ordered segments per bovine chromosome are indicated for these loci in the column S.C. A four digit number appears, i.e., 2222 which means 2 conserved syntenies cattle-human, 2 conserved syntenies cattle-mice, 2 ordered segments cattle-human and 2 ordered segments cattle-mice. The distance in cM from the proximal part of the map is indicated so that 1:1 is chromosome1:1cM. Interesting individual and group comparisons are indicated with superscripts. Gene locations were taken from GDB, MGD, RATMAP and PiGMaP. Locus Bta Hsa Mmu Rno Ssc S.C. IFNAR1: SOD1: 9 KAP8: POU1F1:d 13:116 UMPS: CRYG8: SST:d KNG: CRYAA: PDE1: 3333 SSBP: ACVR2: PDE6A: FN1: INHA: AK2: FCGR1: OSG: IL6:b 1:1 1:3 2222 1:0-8 1:40 21q22.1 21q22.1 3p11 1:66 (q31-q36) 1:90 1:90 (q23-q25) 1:113 3q27 1:139 2:26 16:62.8 16:61.2 11 16:42.7 3q13 3 3q28 21q22.3 2:43-65 UA 2:76-100 UA 2:84-92 2:100-118 2q34 2:115 2q33-q34 2:125 1p34 3:24 1q21 11.. 3:45 4:0-3 7p21-p15 16:19.4 11 17:17.4 UA 5q31.2-q34 18:31.0 1:36.1 1:41.6 9 4 3:49.0 5:17.0 UA 4 UA 1434 DDC:b TCRG:b TCRB:b SYT1: 4:7-17 7p11 4:69-81 4:88-90 5:4 12cen-q21 11:7.0 7p15-p14 13:10.0 7q35 6:19.0 10:38.5 14 17 4 2233 IFNG: 5:32-46 (q22-q24) 12q24.1 5:19 COL2A1:a LYZ: IGF1: 5:75 10:64.0 5:39 12q12-q13.2 15:47.0 5:37-53 (q23) 12 10:63.5 5:49-63 12q22-q23 10:46.0 7 MB: NUBA: CYP2D@: IF:b 5:56 5:94 22q12 15:43.7 EGF:b SPP1: 5:101-117 22q13 15:47.2 6:0-45 4q24-q25 3:87.7 1235 6:36 4q25 3:65.2 6:67 4q11-q21 5:61.0 8:140 KIT:f GABRA2:f FABP-HL: CSN1A: 6:101 4q12 6:104-106 4p13-p12 6:106-108 6:111 (q26-q33) 5:45.2 5:43.0 UA 5:47.0 8:15 CSN3:e 6:111 8:100-110 ALB:f 6:112-126 4q11-q14 5:54.0 8:5.0 (q12) FGFR3:f 6:124-166 4p16.3 5:20.0? QDPR: 6:138-152 4p15.3 5:30.0 PDE6B: 6:145 4p16.3 5:18.84 LDLR: 7:14-50 19p13.2 9:5.0 2535 AMH: 7:32 19p13.3 10:41.5 CSF2: 7:32 5q23-q31 IL4: 7:43 5q23-q31 11:28.0 SPARC: 7:58-96 5q31-q33 11:30.0 FGF1: 7:70-84 5q31.3-q33.2 18:19.0 RASA:b 7:104 (q24-ter) 5q13 13:45.0 Locus Bta Hsa Mmu 8p22 8:32.0 Rno 10 Ssc S.C. CPNB: 8:19 11.. LPL: 14q12-q14 GGTB2: ALDOB: C5: SOD2:b 5 CGA:b GJA1:b CLTA: 11.. TCRA: BRN: SRN: AKAP: ACTG2: CD8A: GGTA1: SHR: 8:117-129 8:129 8:147 8:187 16 9p21-q13 4:18.6 9q22.3-q31 nms 6q25.2 9q33 9:22-34 1212 9:50 6q14-q21 4:9.5 9:90-122 6q21-q23.2 10:26.5 10:0-3 12q23-q24 10:2-18 10:23 10:23 10:82 11:14-54 2222 11:54 2p12 11:103 11:103 17:7.6 14q11.2 14:19.7 2p13.1 UA 6:31.5 9q33-q34 2:26 1:110-120 DBH: LGB: VIM:c 11:138-144 9q34.3 11:141 (q28) 13:44 10p13 2:15.0 2:7.0 2121 CHGB: AVP: GHRH: TG:b IL7:b CRH:b NCAM:b MCAM: PTH:f ARRB: HBB:f KCNA4:f FSHB:f PIGR:e ATP2B4:e 14:51 FH:e ADORA1:e AT3:e FGG: 8:20 FGF2: GUCY: ALDH2: ADRBK2: MT2A: 13:66-72 20pter-p12 2:75.0 13:68-82 20p13 2:73.0 13:96-110 20q11.2 2:87.0 14:1-33 (q12-q16) 8q24 15:35.09 1222 14:37-43 8q12-q13 3:6.6 14:54 8q13 3:8.0 15:26 11q22.2-q22.3 9:28.0 1333 15:35-39 UA 15:51 (q22-q27) 11p15.2-p15.1 7:53.0 15:63 7:50.3 15:63 (q22-q27) 11p15.4 7:50.0 15:69 11p14 2:61.0 15:72 (q24-ter) 11p13 2:60.0 2:37 (p16-p12) 16:5 (q13) 1q31-q41 UA 1111 16:5 1q25-q32 1:70.3 16:5 1q42.1 1:74.1 16:1-9 1q32.1 UA 16:73 1q23-q25.1 1:84.6 17:6 (q12-q23) 4q28 3:46.4 2222 17:51-53 4q25-q27 3:19.9 17:67-71 17:84-110 12q24.2 UA 17:140 22q11 5:66.0 18:24-52 16q13 8:45.0 7 8 1 1q22 2q31-q34 2222 MC1R: PIM1:cg GH1: 18:26-50 16q24.3 8:68.0 18:99 6p21 17:16.4 19:44-58 (q17-qter) 17q22-q24 11:65.0 12:32 (p14) 1111 MAPT: 19:60-70 17q21 11:64.0 GFAP: 19:65 17q21 11:62.0 HD5S39 20:16 5q11.2-q13.3 1232 HEXB: 20:16 5q13 13:50.0 MAP1B: 20:22-28 (q14) 5p13 HTR1A: 20:25 5cen-q11 13:57.5 ANP1: 20:55 GHR: 20:55 5p14-p12 15:4.6 16q13-q14 PRLR: 20:55 5p14-p13 15:4.6 2 Locus Bta S.C. Hsa Mmu Rno Ssc IGF1R: 21:6-24 11.. 22:88 3q11-q12 22:100 3p25 23:5 6q21.3 GPX1: HRH1: DYA: 15q25-qter 7:33.0 UA 4 1212 VEGF: MOG: TNFA: DRB: 23:18 23:30-36 23:30-34 23:32 7:50-60 CYP21: 7 SMHCC: PRL: 7:68 FECH: PACAP: CDH2: DSC3:d CYB5: PRM1: EPO: PLANH: ASAT: 11.. DNTT: PSAP: RBP3: GSTP1: OPCML:d CD5: MAOA:e 23:41 UA UA 6p22-p21.3 6p21.3 6q21.3 23:33 6p21.3 23:33 6p21.3 6p22.2-p22.1 20 17:19.06 17:18-23* 20 20 17:18.77 17:18-23* 13:14.0 24:0-24 18q21.1-q21.31 18:39.0 2122 24:15-33 24:28 18q12.1 18:6.0 24:34 9p 18:7.4 24:63-73 18q22.3-q23 25:16-28(q12-13) 6p13.13 16:3.4 23.. 25:56-62 7q21.3-q22.1 5:84.0 25:69 7q22.1-q22.3 1:61.1 26:35 26:29-41 10q23-q24 19:44.0 28:2-48 10q21-q22 10:27.3 12.. 28:92 10q11.2 14:13.0 29:74 11q13 19:0.0 1212 29:82 11 9:10.0 29:97 11q13.1 19:5.0 X:65 Xp11.4-p11.3 X:5.0 20 17 10q12 12 16 Total S.C. 37 53 31 35 Conserved syntenies cattle-human = 37 cattle-mouse = 53 Ordered segments cattle-human = 31 cattle-mouse = 35 UA: unassaigned blank: no reference found *: MHC region in mice a: Loci close to an evolutionarily conserved breakpoint b: Lack of conservation of synteny cattle-mice coupled to lack of conservation of gene order cattle-humans c: Conservation of synteny of human-mice but not of cattle d: Conservation of gene order or synteny cattle-mice but not for cattle-human e: Lack of conservation of distance between genes or chromosomal landmarks f: Conserved segment with different orders in cattle, humans and mice g: Compare to other loci on BTA23, HSA6p and MMU17 GDB v6.0 11 March 1996 MGD 13 March 1996 PiGMaP 14 March 1996 RATMAP 18 March 1996






![[Answer Sheet] Theoretical Question 2](http://s3.studylib.net/store/data/007403021_1-89bc836a6d5cab10e5fd6b236172420d-300x300.png)


