Problem Set 1, Bio 4181: Due Sept
advertisement

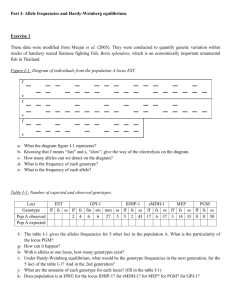
Problem Set 2, Bio 4181: Due Sept. 21, 2011 1. Consider the following pedigree: a). What is the pedigree inbreeding coefficient F of the female shown by the solid circle relative to this pedigree? b). What is the probability of identity by descent in the female shown by the solid circle of a randomly chosen X-linked locus relative to this pedigree? 2. Given the following allele frequencies for an autosomal locus with two alleles (A and a, with p being the frequency of A) and inbreeding coefficients (measured as a deviation from Hardy– Weinberg proportions in all problems in this set), calculate the genotype frequencies. a. p= 0.1, f = 0.6 b. p= 0.25, f = -0.1 c. p= 0.6, f = -0.4 d. p= 0.6, f = 0.4 3. Estimate the value of f for each population given the following genotype numbers. Genotypes AA Aa aa a. 10 70 20 b. 40 20 40 c. 10 30 60 4. Suppose two populations are polymorphic at an autosomal locus with two alleles, A and a. Suppose population 1 has p=0.2 (the frequency of A), and population 2 has p=0.9. Both populations are in Hardy-Weinberg. a. Suppose you sample 400 people from population 1 and 600 people from population 2. What are the expected genotype numbers in your total sample? b. What is the f in your sample? 5. A population has two alleles (A and a) at an autosomal locus with the frequency of A (p) being 0.8. Each genotype has a distinct phenotype. Assuming that generation 1 is in Hardy-Weinberg equilibrium and that generations 2, 3 and 4 are all produced by 100% assortative mating from the previous generation, calculate the genotype frequencies, p, and f for generations 1 through 4. 6. Same question as 5, but now assume that generations 2, 3 and 4 are all produced by 100% disassortative mating from the previous generation.