Single Factor Experiments
advertisement

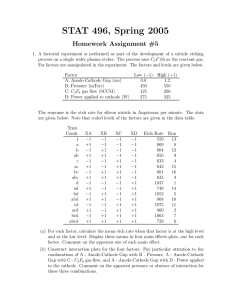
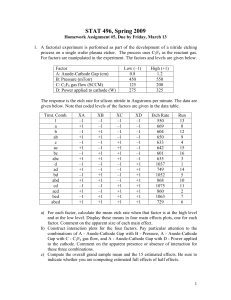
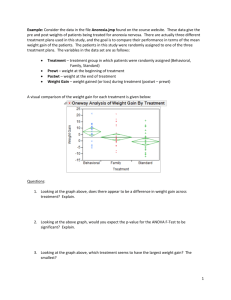
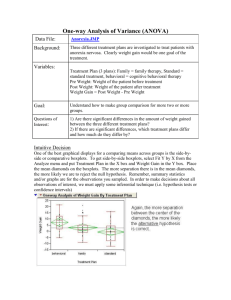
Chapter 3 - Experiments with a Single Factor: The Analysis of Variance (ANOVA) Example 3-1: Plasma Etching Experiment: Power and Etch Rate (pg. 62) Data File: Power-Etch (Ex 3-1).JMP Here are these data entered into JMP. You need two columns, one specifying the Power (W) and the other being the measured etch rate. Make sure Power, although numeric, is interpreted by JMP as a nominal/factor variable. To specify a numeric variable as a nominal/factor variable left-click on the © next Power (W) and change the variable type to nominal as shown below. Analysis in JMP ~ Fit Y by X approach Select Fit Y by X from the Analyze menu and place Power (W) in the X, Factor box and Etch Rate in the Y, Response box, then click OK. From the Oneway Analysis pulldown menu select the following options: Means/Anova Normal Quantile Plots (won't show much here with n = 5 reps) UnEqual Variances (tests whether the population variances are equal) Compare Means > All Pairs, Tukey HSD 1 The resulting output from the Oneway Analysis is shown below: ANOVA Table with Sum of Squares (see pg. 71) H o : 1 2 3 4 or Ho :1 2 3 4 0 H 1 : i j for some i j H 1 ; i 0 for some i p-value < .0001 Reject Ho , at least two mean etch rates differ for the power levels being examined. To determine which power levels significantly differ we need to use am appropriate multiple comparison procedure (see 3.5 pgs. 85-98). CI’s for Treatment Means and Differences in Treatment Means 100(1-)% CI for i (not corrected for experimentwise error rate) y i t / 2, N a SE ( y i ) where SE ( yi ) MS E 8.1695 here. n 100(1-)% CI for ( i j ) (not corrected for experimentwise error rate) ( yi y j ) t / 2, N a SE ( yi y j ) where SE ( yi y j ) 2MS E 11.5534 here. n CI’s for the Mean Etch Rates Under the Different Power Settings (see pg. 74) Model Parameter Estimates: ˆ ˆ1 ˆ2 ˆ3 ˆ4 2 Bonferroni Corrected CI’s If you are going construct r CI’s each at the 95% confidence level the probability that all r of them cover the population parameters of interest is (.95) r 1 r . The probability that least one of the CI’s misses is therefore 1 (.95) r < r . This probability is called the experimentwise error rate. To guarantee that the experimentwise error rate is no larger the specified is simply to use r instead of in the CI formulae on the previous page. This requires that we know in advance how many CI’s we are interested in constructing and that the number of planned (post-hoc) CI’s r is no too large. If r is large we will end up with CI’s that are too wide to be useful. When r is large it is better to use multiple comparison procedures more specific to types of post-hoc comparisons we are interested in doing (e.g. Scheffe’s, Tukey’s, Dunnett’s, etc...) Example: Suppose we wanted to compare all power settings to one another in the etching experiment using 95% confidence intervals for the differences in the 4 mean etch rates. This would require constructing = 6 confidence intervals. 2 So we should use .05 .00833 in place of in the CI’s for the differences r 6 the mean etch rates. Using t-Quantile Calculator we find t.00833/ 2,16 3.34 Comparing Population Variances The text on pgs. 80-82 covers two statistical tests used to assess the equality of the variances across treatment level, Bartlett’s and Levene’s tests. The p-values for both of these tests along with two others (O’Brien & Brown-Forsythe) are reported by JMP when selecting the UnEqual Variances option from the Oneway Analysis pull-down menu. Neither test here suggests inequality of the population variances as both have p-values >> .05. A warning is given because n = 5 replicates limits our ability to detect variance differences. All tests require that the response is normally distributed for each level. H O : 12 22 k2 H 1 : at least two differ 3 Multiple Comparison Procedures Multiple comparison procedures are designed to allow the analyst to make multiple mean comparisons while controlling the experimentwise error rate. Some of the most commonly used multiple comparison procedures are: Tukey’s, Hsu’s, Scheffe’s, and Dunnett’s. A brief description of these procedures is given below: Tukey’s - allows analysts to compare all possible pairs of means Hsu’s – allows the analyst to compare the “best” mean vs. rest. Scheffe’s – allows the analyst to compare all possible linear combinations of means, e.g. the means 1 and 2 contrasted vs. means 3 and 4. Dunnett’s – allows the analyst to compare all means back to a control group or setting. These are all available in some form in JMP, however we will focus our attention on Tukey’s procedure, because of these procedures it is the most widely used. To obtain the results of Tukey’s procedure select Oneway Analysis > Compare Means > All Pairs, Tukey’s HSD (Honest Significant Difference). The results for the etch rate experiment are shown below. Here we see that all power levels significantly differ from each other in terms of the mean etch rate. None of the power settings are connected by the same letter suggesting all are different. It is important to quantify significant differences or effects through the use of CI’s for the pairwise differences in the mean etch rates under the different power settings. The largest difference in etch rate is between the highest and lowest power settings. We estimate the etch rate when the power is set at 220 watts is between 122.75 and 188.85 units larger than the etch rate when the power is 160 watts. Other differences are interpreted in a similar fashion. No of the CI’s contain 0, which supports are conclusion from above, that all power settings result in significantly different mean etch rates. The experimentwise error rate for all comparisons and CI’s is . 4 The Fit Model Approach to Performing a Single Factor ANOVA in JMP As an alternative to the Fit Y by X approach used above we can also use the Analyze > Fit Model option in JMP. There are some advantages to using this approach: It allows for analyzing experiments with more than one factor Allows for checking assumptions using residuals (normality and constant error variance) Allows for checking case diagnostics like outlier-T, Cook’s distance, etc. Gives the estimated parameters for the fitted model. Put the response of interest in the Y box and all factors in the Construct Model Effects box. We will eventually put multiple factors and their interactions in this box. The important aspects of the resulting output are show below ANOVA table gives the usual stuff The parameter estimates are different than those given earlier. Here we have constrained ˆ4 0 so ˆ 617.75 represents the mean for the highest power setting (220 W) rather than the overall mean. This parameterization is required for mathematical reasons which we will not get into at this point. The effect tests section gives the p-value associated with each of the effects in the model. For a single factor experiment it is the same the results from the ANOVA table. The residual by predicted plot shows the residuals ( eij ) plotted vs. the fitted values ( ŷ ij ) ~ i.e. the treatment sample means ( yi ). We want to see no discernible pattern in this plot. If there is fanning out, it can indicated that the variation in the response is not the same across the different treatment levels. 5 To assess the normality of the errors we need to save the residuals or the standardized residuals to the spreadsheet and use Analyze > Distribution to examine their distribution. The distribution of the residuals is slightly kurtotic (S-shape), but not enough to be concerned about. To obtain the results of Tukey’s multiple comparison procedure select that option from the Power (W) pull-down menu to the right of the main section of output. The resulting output includes the connected means report (letter A,B,... one) and you can get CI’s for the pairwise differences in an ordered difference report as in the Fit Y by X case. The matrix below conveys all the same information. 6 Single Factor Experiments in Design-Expert 6.0 First select File > New Design and then choose General Factorial from the Factorial tab. This will open a window where you asked to specify the number of categorical factors you wish to use, which for the etching experiment is one. Next you specify the number of levels for the factor, the name of the factor, the units for factor, and the settings as shown below. Click Continue when you are finished. Next you are asked to enter the number of replicates to use, which in this case is n = 5. Blocking can be specified if necessary. Information about the response(s) is entered next. 7 Data Entry Spreadsheet from Design-Expert Design-Expert will randomize the run orders by default. This is great if we actually going to run the experiment ourselves, but as we already have the data from this experiment it is easier to list the runs in what is referred to as the Standard Order vs. Run Order. To do this click on View > Std Order to change the spreadsheet to standard ordering. We are now ready to conduct our analysis of the data. Begin by clicking the Analysis icon in the menu tree on the left. Design-Expert pretty much leads you by the nose at this point. Click on the name of the response (Etch Rate) to analyze it. In experiments where there are more than one response need to analyze each separately. 8 Choice of Response Transformation (Transform) You are first asked to choose a transformation for the response. Unless you have a prior knowledge about the response that suggests the need for a transformation, we begin our analysis without one. Selecting Model Effects (Effects) Here we choose which effects wewish to examine in the model. The default chosen by Design-Expert is generally recommended, but when we get into multi-factor experiments we may decide to eliminate some effects from the model before proceeding with our analysis. The significance of the chosen effects is also reported here, which can help with effect selection. 9 Analysis of Variance (ANOVA) The ANOVA tab gives a full statistical report for the model we have fit. 10 Case Diagnostics Examining case diagnostics is generally more important we more complicated designs or when regression is being used to predict the response under different experimental conditions. The only case statistic of interest to use at this point would be the Outlier-t which is used to flag outliers. Outliers will have * next to them. Diagnostic Plots Normal Probability (normal probability/quantile plot of the residuals) * Residuals vs. Predicted Values * Residuals vs. Run Order (only if we actually did the experiment in the order) * Residuals vs. Factor (only need to look at this in multi-factor experiments) Outlier T (visualization of the outlier-t statistic results) * Cook’s Distance (visualization of Cook’s influence measure) Leverage (visualization of the potential for influence) Predicted vs. Actual (visualization of how well the model fits the data) Box-Cox (used to find a transformation to improve normality of the response) * In a single factor experiment we really only need to look at the plots denoted by a (*) in the above list. Also we only need to look at the Box-Cox transformation if we see problems, non-normality or heteroscedasticity, in the residual plots. Some these plots are shown below. 11 There is no graphical evidence of the need to transform from either the normal probability plot of the residuals or from residuals plotted vs. the predicted values. If there had been evidence of assumption violations from these plots we would generally examine the results of the Box-Cox procedure to see a suitable transformation can be found. No transformation is recommended here, which is what we expected given the residual diagnostic plots above. Model Graphs For a single factor experiment the standard comparative display is used to visualize results. The raw data is denoted by the red dots and the sample means for each factor setting along with CI’s are also shown in black. We will discuss the Optimization section later when we get into multi-factor experiments and response surfaces. 12 13