Lecture 12-Linkage and Genetic Mapping in Eukaryotes
advertisement

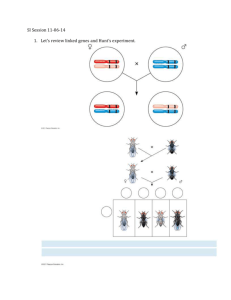
Modified from http://www.mhhe.com/brooker BIO 184 Fall 2006 LECTURE 12 Lecture 12: Linkage and Genetic Mapping in Eukaryotes Map of human chromosome 21 indicating the relative locations of known disease-causing genes. These genes were originally mapped using genetic mapping (linkage mapping) as described in this set of lecture notes. http://www.ornl.gov/sci/techresources/Human_Genome/posters/chromosome/chromo21.shtml The genomes of eukaryotic organisms contain hundreds to thousands of genes (an estimated 30,000-50,000 in humans). Yet there are only a handful of chromosomes. Thus, each chromosome in a eukaryotic genome must contain a large number of genes. The transmission of genes located on the same chromosome may violate Mendel’s Law of Independent Assortment, particularly if they are located very close together along the same arm of a chromosome. This set of lecture notes will explain why, and provide the theoretical basis for mapping genes along a chromosome by following the degree to which they violate Mendel’s Law of Independent Assortment during genetic crosses. Page 1 Modified from http://www.mhhe.com/brooker BIO 184 Fall 2006 LECTURE 12 I. Linkage and Crossing Over In eukaryotic species, each linear chromosome contains a long piece of DNA A typical chromosome contains many hundred or even a few thousand different genes The term “linkage” has two related meanings o 1. Two or more genes can be located on the same chromosome o 2. Genes that are close together tend to be transmitted as a unit Chromosomes are called linkage groups They contain a group of genes that are linked together The number of linkage groups is the number of types of chromosomes of the species o For example, in humans 22 autosomal linkage groups An X chromosome linkage group A Y chromosome linkage group Genes that are far apart on the same chromosome may independently assort from each other due to crossing-over during meiosis. Occurs during prophase I of meiosis Homologous chromosomes exchange DNA segments See Figure 5.1, Brooker In 1905, William Bateson and Reginald Punnett conducted a cross in sweet pea involving two different traits: flower color and pollen shape This is a dihybrid cross that is expected to yield a 9:3:3:1 phenotypic ratio in the F2 generation However, Bateson and Punnett obtained surprising results See Figure 5.2, Brooker They suggested that the transmission of the two traits from the parents was somehow coupled The two traits are not easily assorted in an independent manner Page 2 Modified from http://www.mhhe.com/brooker BIO 184 Fall 2006 LECTURE 12 However, they did not realize that the coupling is due to the linkage of the two genes on the same chromosome The first direct evidence of linkage came from studies of Thomas Hunt Morgan Morgan investigated several traits that followed an X-linked pattern of inheritance Figure 5.3, Brooker, illustrates an experiment involving three traits o Body color o Eye color o Wing length However, Morgan still had to interpret two key observations: 1. Why did the F2 generation have a significant number of nonparental combinations? 2. Why was there a quantitative difference between the various nonparental combinations? Let’s reorganize Morgan’s data by considering the pairs of genes separately: Gray body, red eyes 1,159 Yellow body, white eyes 1,017 Gray body, white eyes Yellow body, red eyes Total 17 12 2,205 Red eyes, normal wings 770 White eyes, miniature wings 716 Red eyes, miniature wings White eyes, normal wings Total 401 318 2,205 Page 3 But this nonparental combination was rare It was fairly common to get this nonparental combination Modified from http://www.mhhe.com/brooker BIO 184 Fall 2006 LECTURE 12 To explain these data, Morgan considered the previous studies of the cytologist F.A Janssens Janssens had observed chiasmata microscopically and proposed that crossing over involves a physical exchange between homologous chromosomes Morgan shrewdly realized that crossing over between homologous X chromosomes was consistent with his data and that: o Crossing over did not occur between the X and Y chromosome o The three genes were not found on the Y chromosome Morgan made three important hypotheses to explain his results: 1. The genes for body color, eye color and wing length are all located on the X-chromosome. Thus, they tend to be inherited together. 2. Due to crossing over, the homologous X chromosomes (in the female) can exchange pieces of chromosomes This created new combination of alleles 3. The likelihood of crossing over depends on the distance between the two genes Crossing over is more likely to occur between two genes that are far apart from each other See Figure 5.4, Brooker II. Genetic Mapping in Plants and Animals Genetic mapping is also known as gene mapping or chromosome mapping. Its purpose is to determine the linear order of linked genes along the same chromosome Figure 5.8, Brooker, illustrates a simplified genetic linkage map of Drosophila melanogaster Genetic maps are useful in many ways: Page 4 Modified from http://www.mhhe.com/brooker BIO 184 Fall 2006 LECTURE 12 1. They allow us to understand the overall complexity and genetic organization of a particular species 2. They improve our understanding of the evolutionary relationships among different species 3. They can be used to diagnose, and perhaps, someday to treat inherited human diseases 4. They can help in predicting the likelihood that a couple will produce children with certain inherited diseases 5. They provide helpful information for improving agriculturally important strains through selective breeding programs Genetic maps allow us to estimate the relative distances between linked genes, based on the likelihood that a crossover will occur between them. Experimentally, the percentage of recombinant offspring is correlated with the distance between the two genes If the genes are far apart many recombinant offspring If the genes are close very few recombinant offspring Map distance = Number of recombinant offspring X 100 Total number of offspring The units of distance are called map units (mu) or centiMorgans (cM). One map unit is equivalent to 1% recombination frequency. Genetic mapping experiments are typically accomplished by carrying out a testcross A mating between an individual that is heterozygous for two or more genes and one that is homozygous recessive for the same genes A. Dihybrid Test Cross Mapping Figure 5.9, Brooker, provides an example of a testcross involving two genes Page 5 Modified from http://www.mhhe.com/brooker BIO 184 Fall 2006 LECTURE 12 o This cross concerns two linked genes affecting bristle length and body color in fruit flies: s = short bristles s+ = normal bristles e = ebony body color e+ = gray body color One parent displays both recessive traits – i.e. it is homozygous recessive for the two genes (ss ee) The other parent is heterozygous for the two genes The s and e alleles are linked on one chromosome The s+ and e+ alleles are linked on the same homologous chromosome The data at the bottom of Figure 5.9 can be used to estimate the distance between the two genes: Number of recombinant offspring X 100 Total number of offspring 76 + 75 X 100 542 + 537 + 76 + 75 = = 12.3 map units (cM) Therefore, the s and e genes are 12.3 cM apart from each other along the same chromosome. B. Trihybrid Test Cross Mapping Data from trihybrid crosses can also yield information about map distance and gene order The following experiment outlines a common strategy for using trihybrid crosses to map genes In this example, we will consider fruit flies that differ in body color, eye color and wing shape. Page 6 Modified from http://www.mhhe.com/brooker BIO 184 b = black body color b+ = gray body color pr = purple eye color pr+ = red eye color vg = vestigial wings vg+ = normal wings Fall 2006 LECTURE 12 Step 1: Cross two true-breeding strains that differ with regard to three alleles. Male is homozygous wildtype for all three traits Female is mutant for all three traits The goal in this step is to obtain F1 individuals that are heterozygous for all three genes Step 2: Perform a testcross by mating F1 female heterozygotes to male flies that are homozygous recessive for all three alleles Page 7 Modified from http://www.mhhe.com/brooker BIO 184 Fall 2006 LECTURE 12 (Produced by cross above) During gametogenesis in the heterozygous female F1 flies, crossovers may produce new combinations of the 3 alleles. Step 3: Collect data for the F2 generation Phenotype Number of Observed Offspring Gray body, red eyes, normal wings 411 Gray body, red eyes, vestigial wings 61 Gray body, purple eyes, normal wings 2 Gray body, purple eyes, vestigial wings 30 Black body, red eyes, normal wings 28 Black body, red eyes, vestigial wings 1 Black body, purple eyes, normal wings 60 Black body, purple eyes, vestigial wings 412 Analysis of the F2 generation flies will allow us to map the three genes The three genes exist as two alleles each Therefore, there are 23 = 8 possible combinations of offspring If the genes assorted independently, all eight combinations would occur in equal proportions Page 8 Modified from http://www.mhhe.com/brooker BIO 184 Fall 2006 LECTURE 12 It is obvious that they are far from equal In the offspring of crosses involving linked genes: o Parental phenotypes occur most frequently o Double crossover phenotypes occur least frequently o Single crossover phenotypes occur with “intermediate” frequency The combination of traits in the double crossover tells us which gene is in the middle o A double crossover separates the gene in the middle from the other two genes at either end In the double crossover categories, the recessive purple eye color is separated from the other two recessive alleles o Thus, the gene for eye color lies between the genes for body color and wing shape Step 4: Calculate the map distance between pairs of genes To do this, one strategy is to regroup the data according to pairs of genes: From the parental generation, we know that the dominant alleles are linked, as are the recessive alleles This allows us to group pairs of genes into parental and nonparental combinations Page 9 Modified from http://www.mhhe.com/brooker BIO 184 Fall 2006 LECTURE 12 Parentals have a pair of dominant or a pair of recessive alleles Nonparentals have one dominant and one recessive allele The regrouped data will allow us to calculate the map distance between the two genes Parental offspring Total Nonparental Offspring Total Gray body, red eyes (411 + 61) 472 Gray body, purple eyes (30 + 2) 32 Black body, purple eyes (412 + 60) 472 Black body, red eyes (28 + 1) 944 61 29 The map distance between body color and eye color is therefore: 61 X 100 = 6.1 cM 944 + 61 Map distance = Parental offspring Total Nonparental Offspring Total Gray body, normal wings (411 + 2) 413 Gray body, vestigial wings (30 + 61) 91 Black body, vestigial wings (412 + 1) Black body, normal wings (28 + 60) 88 826 413 179 And the map distance between body color and wing shape is: Map distance = 179 X 100 = 17.8 cM 826 + 179 Page 10 Modified from http://www.mhhe.com/brooker BIO 184 Parental offspring Fall 2006 LECTURE 12 Total Nonparental Offspring Total Red eyes, normal wings (411 + 28) 439 Red eyes, vestigial wings (61 + 1) 62 Purple eyes, vestigial wings (412 + 30) Purple eyes, normal wings (60 + 2) 442 881 62 124 And, finally, the map distance between eye color and wing shape is Map distance = 124 881 + 124 X 100 = 12.3 cM Step 5: Construct the map Based on the map unit calculation, the body color and wing shape genes are farthest apart and the eye color gene is in the middle: ? Page 11