GENE EXPRESSION BACKGROUND INFORMATION SHEET
advertisement

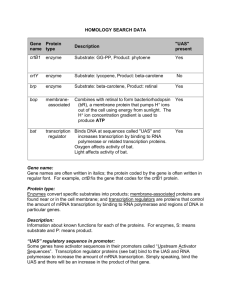
HOMOLOGY BACKGROUND INFORMATION Your goal is to determine relationships between proteins that define the cellular network. In order to do this your group will analyze data obtained from homology searches. Scientists are working to determine relationships between proteins by compiling all available information about the genes and proteins in a network into a very large database. The gene sequences for genomes of many organisms have been added to this database. A genome is an organism’s entire sequence of DNA; therefore, if the DNA sequence is known, the proteins can be predicted (recall DNA mRNA protein). For many genes, functions and properties have been determined in the laboratory. But for many other genes and proteins, scientists use this database to help them make hypotheses about the functions and properties of these genes and proteins. Scientists compile what is known about the genes and proteins in the network of interest. Then they analyze the information to hypothesize relationships between the proteins. Information about genes with unknown functions can be predicted by comparing their sequences to genes in the database that have known functions and properties. Searching for homologous genes, genes with similar sequences, is called homology searching. Similar nucleotide sequences will result in proteins that have similar shapes and folding patterns. Homologous proteins that are similar in their 3-dimensional shape will have similar properties and functions. Homologous proteins are proteins that perform the same function in two different organisms. For example, the hemoglobin protein that carries oxygen in the blood of a cow is homologous to the hemoglobin protein in humans. Information obtained from homology searching can help scientists determine if the protein they are studying is an enzyme, a membrane-associated protein, or a transcription regulator. Scientists can then study the protein’s role in the network more specifically. Scientists can also determine what kind of regulatory sequences exist in the DNA that will affect the transcription of the gene that codes for the protein. Ultimately, all this information helps scientists determine how a protein interacts in a metabolic network. HOMOLOGY SEARCH DATA Gene name Protein type Description "UAS" present crtB1 enzyme Substrate: GG-PP, Product: phytoene Yes crtY enzyme Substrate: lycopene, Product: beta-carotene No brp enzyme Substrate: beta-carotene, Product: retinal Yes bop membraneassociated Combines with retinal to form bacteriorhodopsin (bR), a membrane protein that pumps H+ ions out of the cell using energy from sunlight. The H+ ion concentration gradient is used to produce ATP Yes bat transcription regulator Binds DNA at sequences called "UAS" and increases transcription by binding to RNA polymerase or related transcription proteins. Oxygen affects activity of bat. Light affects activity of bat. Yes Gene name: Gene names are often written in italics; the protein coded by the gene is often written in regular font. For example, crtB1is the gene that codes for the crtB1 protein. Protein type: Enzymes convert specific substrates into products; membrane-associated proteins are found near or in the cell membrane; and transcription regulators are proteins that control the amount of mRNA transcription by binding to RNA polymerase and regions of DNA in particular genes. Description: Information about known functions for each of the proteins. For enzymes, S: means substrate and P: means product. “UAS” regulatory sequence in promoter: Some genes have activator sequences in their promoters called “Upstream Activator Sequences”. Transcription regulator proteins (see bat) bind to the UAS and RNA polymerase to increase the amount of mRNA transcription. Simply speaking, bind the UAS and there will be an increase in the product of that gene. ANALYSIS: 1. Fill in the spaces below using the information provided on the Homology Data Table. Fill in the gene name (protein) that catalyzes each reaction. lycopene Protein A T P GG-PP beta-carotene phytoene Protein Protein retinal phytoene brpProtein bacteriorhodopsin 2. What is the result of the UAS being bound? 3. In the production of bacteriorhodopsin which protein binds to UAS? 4. If the transcription regulator is bound to the UAS on the DNA, which genes will have their products increase (see table)? 5. In the presence of light, bat production will increase. Knowing this, how does light effect the pathway of bacteriorhodopsin production? Use the diagram above to help explain the steps involved. KEY FOR HOMOLOGY DATA Directions: Fill in the gene name (protein) that catalyzes each reaction using the information provided on the Homology Data Table. Your goal is to determine how light affects the amount of bacteriorhodopsin in the halo cell. lycopene P r crtY o Protein t e beta-carotene i n GG-PP crtB1 ProteinProtein Bop retinal ATP Protein bacteriorhodopsin 1. How does the amount of light affect the amount of bacteriorhodopsin in halo? Increase light = increase bacteriorhodopsin 2. Using your data table as a guide, which protein is likely the sensor that detects the presence of light and oxygen? What specific information suggests this? bat-- Binds DNA at sequences called "UAS" and increases transcription by binding to RNA polymerase or related transcription proteins. Oxygen = more likely to bind at UAS = more transcription Oxygen affects bat’s ability to bind UAS Light affects bat’s ability to bind UAS. 3. What does UAS mean? Which genes have a UAS? How does having a UAS affect the amount of transcription of the genes? “UAS” regulatory sequence in promoter: Some genes have activator sequences in their promoters called “Upstream Activator Sequences”. Transcription regulator proteins bind to the UAS and RNA polymerase to increase the amount of mRNA transcription. 4. Which protein binds to the UAS? bat 5. Thinking of your responses to the above questions, suggest a hypothesis for how the network is regulated in response to light and oxygen. You can explain your hypothesis with a diagram in addition to words. Responses will vary 6. What data/information would help you test the hypotheses or support or refute the hypothesis you suggested in question #5? How the biochemistry is involved; what happens if a piece is missing (gene expression).