Supplementary Methods and Figure Legends -
advertisement

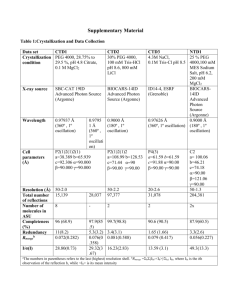
1 Supplementary Information Supplementary Methods Cloning, Expression and Purification All constructs were generated using a standard PCR-based cloning strategy, and entire coding sequences were verified by sequencing. The whole genomic DNA for geminin (residues 1–206) and Cdt1 (residues 1-557) gene were cloned from mouse embryonic fibroblast cDNA, inserted into pGEX-4T3 vector, and expressed in Escherichia coli BL21 as GST fusion proteins. Each protein was purified first by glutathion sepharose column followed by cleavage of GST with thrombin. Full length geminin was further purified using anion exchange (Mono-Q) and gel-filtration chromatography (Superdex 75 column). Cdt1 was further purified by cation exchange (Mono-S) and gel-filtration chromatography. To define the domain boundaries in Cdt1 and geminin, we employed an approach that combined limited proteolysis using subtilisin, sequence alignment with other Cdt1 or geminin family members (Supplementary Fig. 2), and binding assays between the Cdt1 and geminin fragments (Supplementary Table 2). This characterization identified a conserved core fragment for Cdt1 (tCdt1, residues 172-368) and Geminin (tGeminin, residues 79-157). His-tagged tGeminin and tCdt1 were also synthesized with a PCR based method. Both PCR products were digested with NdeI-XhoI, and inserted into a modified PETDuet vector (tCdt1) and a pACYC vector (tGeminin, Novagen). The plasmids containing the tGeminin gene and the tCdt1 gene were co-transformed into E. coli BL21 (DE3), and the bacteria were grown in LB media containing 50 µg/ml Kanamycin and 34 ug/ml Chloramphenicol until an A600 of 0.6 was reached before induction with 0.4 mM isopropyl-1-thio-β-D-galactopyranoside for 4 h. Initially, the protein 2 complexes were purified by nickel column using a 20-250 mM imidazole gradient in 50 mM NaH2PO4, 300 mM NaCl, and 5 mM 2-mercaptoethanol, pH 8.0. The eluted proteins were further purified with a 0-500 mM NaCl gradient on a Mono-Q column, and finally, by gel filtration on a Superdex 75 column equilibrated with 50 mM BTPHCl, 200 mM NaCl, 5 mM DTT, pH 6.8. The tGeminin-tCdt1 complex used for crystallization was concentrated to 8–10 mg/ml, and proteins for biochemical assays were concentrated to 0.5–2 mg/ml. Selenomethionine (Se-Met)-substituted protein complex was cultured by growing the plasmids in E. coli strain B834 (DE3) in M9 minimal medium. Mutagenesis All the geminin and Cdt1 mutants used in this study were created by PCRbased methods (Stratagene). The mutant proteins were purified using affinity chromatography followed by Mono-S (or Mono-Q for geminin mutants) and gel filtration chromatography, as described above. Five residues (Glu113, Asn114, Glu115, His118, Lys119) and seven residues (Leu107, Leu111, Asn114, Glu115, His118, Lys119) were simultaneously mutated to alanine in tGemininmis5 and tGemininmis7, respectively. Eight residues (Lys133Ser, Lys136Ser, Asp137Ser, Glu140Ala, Glu143Ala, Gln146Ser, Tyr147Ala, Met148Ala) were simultaneously mutated in tGemininmis8. We designed tGeminin linker mutants with a flexible linker of 6 or 12 residues (KGSREG or KGTREGKGSPEG) inserted between residue 135 and 136, or replaced the residues 136-141 or 136-147, respectively. The chimeric tGemininGCN4 mutant was created by fusing Saccharomyces cerevisiae GCN4 (residues 251–262, KQLEDKKVEELLS) between Asn135 and Met148 of tGeminin. Circular dichroism analysis and proteolytic digestion confirm that the inserted residues form a flexible linker or a continuous helix (tGemininGCN4). These mutants retain similar binding affinity to Cdt1 (Supplementary Table 2) and similar stability and conformation 3 compared to those of wild-type tGeminin (Supplementary Fig. 4). The flexible linker mutants in which a linker was inserted or replaced behaved similarly in our assays, and we thereafter focus on to the mutants with a 6 residues (tGemininlink6) and a 12 residues (tGemininlink12) inserted linker. Circular dichroism analysis Structural changes of various tCdt1 and tGeminin proteins (30 uM) were monitored by a circular dichroism (CD) spectrometer (Jasco J-715) at various wavelengths (200-250 nm). Conformational changes for each mutant were also monitored at various temperatures by scanning with the CD spectrometer. The temperature was raised from 5 to 85 °C over a 65-min period with detection was set on 222 nm. All the samples used here were prepared in 20 mM KH2PO4, 150 mM NaCl, pH 7.4. DNA-cellulose binding assay 20 ug of His-tagged tCdt1 or tCdt1184-368 protein was incubated with 100 ug of tGeminin (or geminin89-157) and then single stranded DNA-cellulose (Sigma) in 10mM Tris-HCl, 5mM DTT, pH 7.5, for 3hrs at 4℃. After washing with 50mM Tris-HCl, 50mM KCl, 10mM MgCl2, 5mM DTT, pH 7.5, bound proteins were eluted with the same buffer containing 300mM NaCl. Proteins were subjected to SDS-PAGE followed by Western blot analysis using an anti-His-tag antibody. 4 Supplementary Figure Legends Supplementary Figure 1. Heptad repeat and intermolecular interactions within tGeminin dimer. a, A helical wheel representation of the repeated sequence of the tGeminin coiled-coil highlighting the [a] and [d] positions at the center of the dimer interface. The positions of the heptad repeat are labeled [a]–[g]. The coiled-coil dimer of tGeminin shows a classical ‘knobs into holes’ arrangement, where the [a] and [d] positions in the six heptad repeats are populated by eight non-polar residues and four polar residues. Residues that form ion-pairs are connected by dotted lines. The ion-pairs shown in the C-terminal region of the Hb helix are bent and the residues in this region (139–156) are not involved in the dimerization packing in our structure. The Cα atoms of residues 97– 138 of each monomer can be superimposed with an r.m.s. deviation of 0.77 Å. b, Details of the tGeminin dimer. Two different views of the dimerization interaction are shown. Dotted lines indicate hydrogen bonds (Glu113–Asn114, Asn114–Ala110, Asn135–Asn135, Arg103–Ala100) and four ion pairs. Supplementary Figure 2. Structural alignment and conservation in geminin and Cdt1. a, Sequence identity between mouse (Mm) geminin and its orthologues from human (Hs), frog (Xl), fish (Ol) and fly (Dm). Green dots and purple dots indicate tGeminin residues in the Ha and Hb helices, respectively, that contact tCdt1. Every tenth residue is marked with a star. The heptad repeats are boxed with a thick line. b, Secondary structure of mouse (Mm) Cdt1 showing identity with orthologues from human (Hs), frog (Xl), fly (Dm) and fission yeast (Sp). Orange dots and purple dots 5 indicate residues that contact geminin in first and second interface, respectively. Every tenth residue is marked with a star. Supplementary Figure 3. Interaction between tCdt1 and naked DNA. a, Both tCdt1 and Cdt1184-368 proteins associated with DNA, and tGeminin inhibit the DNA binding activity of tCdt1 proteins, supporting the previous observation19. Geminin89-157, however, did not efficiently inhibit the tCdt1-naked DNA binding, suggesting that the helix H6 and a L2 loop of Cdt1 is important in recognition of DNA. b, Regions of Cdt1 (helices H1 and H2 and a L1 loop, and a helix H6 and a L2 loop) in which DNA is expected to interact is labelled (left) and coloured in yellow (right). Overall structure of tCdt1 is displayed with a backbone diagram (left) and a surface representation (right). tGeminin is shown in green colour (right). Supplementary Figure 4. Structure and stability of various tGeminin and tCdt1 mutant proteins. a, analysis of conformational differences between the wild-type and mutant tCdt1 proteins by circular dichroism (CD). CD spectroscopy measurement showed that all tCdt1 deletion mutants displayed identical patterns of spectra to that of wild-type tCdt1, indicating that no significant changes occurred in the structure of tCdt1upon mutation. b, thermal melting curves of wild type and mutant tCdt1 proteins determined by circular dichroism. The unit (°C) is omitted for clarity. 6 c, analysis of conformational differences between the wild-type and mutant tGeminin proteins by CD. All other mutant tGeminin proteins showed the similar profile as that of the wild type and were omitted from this diagram for clarity. d, thermal melting curves of wild type and mutant tGeminin proteins determined by CD. e, tGeminin and tCdt1190-368 do not form a detectable complex in solution. The tGeminin-tCdt1 and tGeminin-tCdt1190-368 complexes were incubated in 50 mM TrisHCl, 200 mM NaCl, 5 mM DTT, pH 7.5 at 4°C for 60 min before being loaded on a Superdex-200 column. The molecular masses and positions of standards are indicated. Because of the elongated shape of tGeminin, it is eluted much earlier than tCdt1190-368 (Supplementary Table 3).