Chapter 6 Complex traits in plants and animall
advertisement

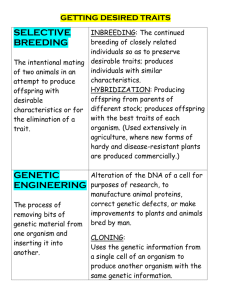
Chapter 6 Complex traits in plants and animalsObjective One of the most challenging problems in genetics is to understand the basis for variation among human beings. It is now widely accepted that the classic question of nature vs. nurture is ill-posed from the start, because there is no simple dichotomy between the two. Traits are not simply genetically based or environmentally based, nor are they a simple addition of genetic and environmental components. Rather the phenotypes that we see are the result of interactions of particular genotypes developing in particular environments. This does not mean that prediction of phenotypes based only on genotypic information is impossible, but it does make the task much more difficult. In this chapter we will explore genetic and phenotypic aspects of human variation, and see what are the prospects for linking the two. Mendelian variation The first kind of variation to consider is the obviously pathological variation caused by single gene defects. Cystic fibrosis is the most common autosomal recessive disorder in the US, and others include PKU, Tay-Sachs disease, and Lesch-Nyhan syndrome. Many of these are defects in metabolism, such that lack of a particular enzyme results in a diseased phenotype. Every one of these births brings heart-breaking news to the parents, and progress in diagnosis, care, and prevention are all proceeding at a rapid rate. Victor McKusick is the editor of an incredible compendium called Mendelian Inheritance in Man which lists over 5000 single-gene disorders. The on-line version of this book is well worth your taking some time to examine (www3.ncbi.nlm.nih.gov/omim/). Try looking up cystic fibrosis, PKU, or any other single-gene genetic disorder, and you will find that the various databases including Genbank, Medline abstracts, and genetic maps are all linked together. The combined power of genomic-scale sequencing and these information technologies means that we will have mapped and sequenced all such single-gene disorders within a few years. This means that each will be diagnosable pre-natally, and the risk that each couple faces in producing an affected child will be possible to determine. Even single-gene disorders are complicated When new cases of PKU are identified, they are usually referred to genetic counselors and it has become clear that there are varying severities of the disease. Some are relatively easily treated by dietary restriction of phenylalanine, while others require more careful monitoring. In order to determine this, the phenylalanine gene of affected individuals is often sequenced. In this way we have learned that there are over 250 different defects in the gene for PAH, and that most individuals who are affected are not actually homozygotes, but rather are heterozygous for two different defective alleles. Similarly, more than 500 alleles of CFTR, the gene responsible for cystic fibrosis, have been identified. Practically everything that can go wrong with these genes has gone wrong in these cases. There are premature stop codons, incorrect amino acids, intron splicing defects, overexpressing alleles, and underexpressing alleles. Although these alleles segregate like a single Mendelian gene, the diversity of alleles that cause ill-health means that it is often not easy to design a simple test to tell whether an individual has a defective allele. Complex diseases Although there are many genes whose defects cause disorders, each such disorder is quite rare, and even the combined incidence of all single-gene disorders is well under 1% of the population. The bulk of human suffering from health problems arises from diseases like cancer and heart disease. We know that these diseases are not simple one-locus genetic diseases, because when we look at families with affected individuals, we cannot draw pedigrees that explain the transmission of the disease from parent to offspring in a simple Mendelian manner. Nevertheless, we suspect that these diseases have a genetic component because the traits tend to aggregate in families. By this we mean that some families seem to be remarkably free of the disease, and others tend to have far too many cases. Recalling our study of the Luria-Delbrück experiment, there is a jackpot distribution of complex diseases across families which does not fit a Poisson distribution of cases. Such familial aggregation does not guarantee that the disease has a genetic basis, however, because families also share environments, like diet and tendencies to exercise. The problem of identifying genes that affect the risk of disease when there may be many such genes for a given disease, and when their influence on disease risk depends on environmental conditions, is indeed a difficult one. The problem has a long history, however, mostly in the context of crop and domestic animal breeding. The study of quantitative genetics considers the transmission of phenotypes through a model of assumed underlying genes in a sophisticated statistical setting. Before we can return to the issues of human variation, we first need to take a careful look at some of the theoretical and empirical underpinnings of quantitative genetics. Artificial selection One of the best ways to understand quantitative genetics is to examine what happens to a population when artificial selection is applied. Artificial selection takes up a large portion of Darwin's writings, as it demonstrated to him that populations can be made to change dramatically by repeatedly choosing a subset of the population to be parents for the next generation. He displayed the differences among dog breeds as evidence that heritable variation can result in dramatic morphological changes. Today we have access to a huge array of DNA markers which serve to tag variation throughout the genome. Following these markers in the face of artificial selection allows us to track which parts of the genome are responding to the selection, which in turn tells us which genes are responsible for the differences. Before getting into the DNA markers, let's first back up and take a look at a classical experiment in artificial selection. In the early 1900's selection was initiated to increase and decrease the oil content of corn kernels. The figure shows the oil content of the kernels over time, and shows that there are kernels whose oil content is well above that of any natural corn. How can such selection result in phenotypes that are totally outside the range of the initial population? One possibility is that new mutations have arisen during the course of the selection. For such large and long term experiments, this is fairly likely. But even without new mutations, extreme phenotypes may be found by selection putting together combinations of genes that are so improbable in the founding population that they simply were not seen. Pure-breeding lines and sources of variation We need to see the results of one more experiment before we can start to put together a model for quantitative genetic variation. In the early 1900's Johannsen did many crosses with the Princess bean, and he found that by self pollinating the beans over many generations he was able to generate lines that had very little variability and appeared to be stable. He called these pure-breeding lines because the offspring always resembled the parents. Despite the fact that he called them pure-breeding, the bean weights did actually vary within each line. Today we can think of the pure-breeding lines as being completely homozygous, and the variation that is seen in the phenotypes of these lines is due to environmental causes. Johannsen crossed two such pure- breeding lines that differed in bean weight. The parental lines might be AABBCCDD × aabbccdd, so the F1 would be AaBbCcDd. In other words, the F1 are heterozygous at many loci, but all the F1 plants have the same genotype. Thus the variation in phenotype from one F1 plant to the next is entirely environmental in cause, and we should expect the F1 to have about the same variation in bean weight as either parent. This is what Johannsen saw. But when the F1 were selfed, the F2 now display all combinations of genotypes from AABBCCDD, to AaBBCcDD, to aabbccdd, and everything in between. We know that the genotypes must be much more variable, so it is not surprising that the phenotypes also display a much broader variation than either the parentals or the F1. This same approach is still used to reveal the underlying genetic basis of quantitative traits in a wide range of economically important plants. For example few days ago, Dr. Steve Taksley from Cornell University explained how his research program on the genetics of domestication in tomato began with an experiment just like this one. He crossed a large-fruited “big boy” tomato with a small fruited wild variety. The resulting offspring were self fertilized to create an F2 population that was highly variable for fruit size. He was able to use a very clever method of genome mapping to quickly find one of the genes responsible for this variation, a gene he called ORFX, that appears to regulate cell division in tomato. A distant relative of this gene was better known in humans, where it is called a RAS protein gene. Defects in the RAS protein gene are implicated in some cancers (ie, cell division gone awry). Genetic basis for continuous variation To be more explicit about the genetic model for Johannsen's beans, or Tanksley’s tomatoes, consider the cross of the F1 x F1. If there are two genes that affect bean weight, and the effect of genotypes aa, Aa, and AA on weight are to add 0, 1, and 2 grams (and likewise for the B gene), then the distribution of weights of the F2 population will be in the proportions 1 : 4 : 6 : 4 : 1, having phenotypes 0, 1, 2, 3, 4 respectively. This distribution turns out to be binomial. For three genes the phenotypes will have proportions 1 : 6 : 15 : 20 : 15 : 6 : 1 for phenotypes 0, 1, 2, 3, 4, 5, 6. As more genes are added, the closer this distribution comes to the normal or bell-shaped distribution. Parent-offspring regression and heritability A method that has been used to explore the degree to which parents pass on traits to their offspring is the parent-offspring regression. This method was invented by Francis Galton, who happens to have been Charles Darwin's first cousin. The method starts by collecting a set of families with both parents and the offspring. Measurements of the trait are taken for all family members. For each family, you calculate the average of the two parents' values, called the mid-parental value, and you calculate the average of the offspring. On a pair of axes, plot a point for each family where the x value is the midparental value and the y value is the offspring mean. If the offspring are totally random in their phenotype, and have no resemblance to the parents, then these points will be a complete scatter. In this case the best fitting line through the points is said to have a slope of zero. On the other hand, if the largest (or whatever) parents have the largest offspring, then the points will tend to show some degree of a trend, like the figure. The slope of the best fitting line through these points is called the heritability. If the slope is one, the offspring are identical to the parents, and we say that the heritability = 1. Most traits like height have a heritability somewhere in the range of 0.1 to 0.5. Realized heritability Now we can use the parent-offspring regression figure to explain the results of artificial selection experiments. When a subset of the parents are selected each generation, we saw that the mean of the progeny population shifts in the direction of selection, but it does not shift in one generation all the way to the mean of the selected parents. The realized heritability is in fact the fraction of the way from no change to the selected parental mean that the offspring mean changes. To relate this to the idea of a parent-offspring regression, just consider the figure once again. First we normalize the population mean to zero. Let the mean of the selected parents be written as S and the mean of the offspring be R. You can see from the figure that the slope of the regression line is R/S. Because this is the heritability, we can write h2 = R/S, which we can re-write to get R = h2S. This is a prediction equation, which says that the response to selection R is the heritability times the difference between the population mean and the mean of the group that is selected to be parents. An interesting paradox of heritability, which actually makes a lot of sense after some thought, is that traits that seem to be very important to the fitness of the organism, such as size and weight of plant seeds, age at first reproduction, or fundamental aspects of the body plan, have much lower heritability than “trivial” traits such as the size of earlobes, color of hair, and height. In other words, “trivial traits” will often respond better to selection to shift their value than “important traits” will. This is explainable because natural selection works hard to limit genetic variation (and thus heritability) of really important fitness-related traits, while selection is weak on traits that are not related to fitness. Thus, more heritable variation has accumulated for the less important traits. A model for quantitative genetic traits It is important to reiterate that quantitative genetics depends heavily on the construction of a model that tries to describe the relationships among relatives in terms of unobserved genetic variation. The basic assumption of the model is that there are many genes that determine each trait, and that each gene has a small effect. Furthermore the simplest model assumes that the effects are additive over loci, and that genetic and environmental effects simply add. We know from many experimental results that the simple addition of genetic and environmental effects is actually not commonly observed, so a slightly more complicated model incorporates the gene x environment interactions. The range of phenotypes that is observed when a particular genotype is reared in a variety of environments is called the norm of reaction. Norm-of-reaction plots show that some genotypes are well buffered against environmental change, so that the phenotype is relatively constant, but other genotypes produce a wide range of phenotypes when reared in different environments. These concepts are virtually universal in laboratory and farm plants and animals. In humans, identical twins provide us with an opportunity to see the variation among phenotypes that result when the same genotype is reared in different environments. How are genes that cause complex diseases in humans found? Now we are ready to consider how genes that cause complex traits are identified in human families. There are basically four ways that are being applied, and it is important to realize that these methods are still being developed and improved. The first is to obtain large, multi-generation families with affected individuals and to type many genetic markers in the families. Co-segregation of the trait with markers indicates that there is linkage between the marker and an unseen gene that may cause the trait. The second method is to again identify many families that have affected individuals, but to focus on families with pairs of affected siblings. When many genetic markers are scored in these families, markers that show an excess of sharing among affected sibs are likely to be close to the gene or genes that affect the trait. The third method is to simply take large random samples from the population, including both affected and unaffected individuals, and to look for genetic markers that are correlated with the trait. This sounds like a brute force kind of approach, but it has been quite effective in mapping the gene for myotonic dystrophy. The fourth method actually starts with experiments in model organisms, like mice, and entails identification of genes responsible for a related phenotype in the model. Molecular techniques are then used to map and clone the human gene(s). Complications There are four factors that make the identification of genes for complex traits very difficult in humans. The first is that many of the traits involve genes that show incomplete penetrance. This means that an individual may be homozygous for an affected allele, yet they show no aberrant phenotype because the effect of the gene is small enough that other factors compensate. The second complication is that some environmental agents can produce phenotypes that mimic the phenotype of a genetic defect, so called phenocopies. The third complication is that some complex traits can be caused by homozygosity for any of several different genes. Hereditary deafness, for example, has several distinct genetic causes, so different families have quite different forms. Offspring of two deaf parents often have normal hearing as a result of the complementation of the two different defective genes. The fourth complication is that complex traits often have a strong environmental component, and often it is extremely difficult to measure and control the environment. There has been some interest in the genetics of intelligence, for example, and these studies are difficult to interpret because it has proven almost impossible to identify environmental components and to measure their effects in a way separable from genetic effects. Candidate genes The method of candidate genes has a lot of promise for traits where we have some clue about the biochemical basis. In the case of cardiovascular disease, we know, for example, that anything that increases the level of fats and cholesterol in the blood tends to accelerate the rate of formation of atherosclerotic plaques, which in turn cause disease. So any gene that upsets the balance of fats in the blood is a suspect. One such gene is lipoprotein lipase (LPL), which regulates the uptake of lipids from the blood into cells. A rare disorder occurs in individuals who lack this enzyme due to a severe defect in the gene. Individuals with LPL deficiency have hypertriglyceridemia, or too much fat in the blood. Charles Sing, Deborah Nickerson and colleagues recently sequenced LPL in 71 individuals and found that 88 different nucleotide positions varied in this sample. If you were to pick two copies of the gene and compare the two at a single nucleotide position, the chance that they would be different is 0.002. This means the two copies of the gene differ about once every 500 nucleotides. The sequence data allowed the investigators to make many inferences about the age of the variation, the degree of recombination within the gene, and the degree of relatedness of different alleles. Work is in progress in this exciting study to try to associate subtle variation in the sequence of LPL with variation in heart disease risk. This is being done by comparing the LPL sequences in a sample of nearly 3000 people who have already had many physiological parameters for lipid balance measured. It is not clear just to what extent we will be able to predict a person's risk of a complex disease by analysis of DNA sequence differences, but this is certainly an area of very active research. Summary 1. The variance in many important traits is in part caused by variation in multiple genes. 2. Heritability, defined as the fraction of the total variance in a trait that is additive genetic variance, can be estimated from parent-offspring regression. 3. Selection response can be predicted from heritability. Heritability is often lower for fitness related traits than for ‘trivial traits’ that do not influence fitness. 4. Phenotypic variation is not simply the sum of genetic and environmental effects. Gene x environment interaction means the phenotypic effect of some environmental changes depends on the genotype.