Protocol for Serratia marcescens
advertisement

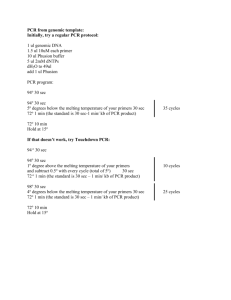
Serratia marcescens--all strains Sample Preparation Because of the potential for PCR inhibitors in host tissue, isolate to improve detection. Dissect vascular bundles from a lower stem section scrubbed with 70% ethanol, place in a 1.5 ml epitube containing 250 l Nanopure water, crush with a sterile golf tee, vortex, and let sit for 30+ minutes (as long as 24 h). Mix suspension and streak onto LB agar or nutrient agar and incubate for 48 hours. Suspend bacterial growth in 1-2 ml Nanopure water; if the cell suspension is completely opaque, dilute until some translucency is achieved. In most instances, a 1/10 dilution is sufficient; some samples need as much as 1/1000 dilution. Reaction Mixture (per 25 l tube) Reaction master mix may be prepared in advance, in darkened conditions, and stored at -18ºC without loss of sensitivity for up to 4 d (the longest intervals tested thus far). 6 l Nanopure water 5 l Smart Cycler Additive Reagent 2.5 l of 2 mM dNTP=s 2.5 l primer YV1 2.5 l primer YV4 2.5 l 10X buffer 0.5 l Titanium Taq polymerase (=0.5 units) 2.5 l of fresh 5X SYBR Green Sample 1.0 l of sample (or controls as noted below) Primers Prepare to a concentration of 0.8 M (0.8 pmol/l) YV1 5'-GGG AGC TTG CTC CCC GG-3' YV4 5'-AAC GTC AAT TGA TGA ACG TAT TAA GT-3' Controls Negative control: Positive control #1: Positive control #2: 1 l Nanopure water 1 l of 100 pg/l of genomic DNA of S. marcescens 102 or 103. 1 l of 100 pg/l of genomic DNA of S. marcescens 102 or 103 plus 1 l of sample. If results show evidence of PCR inhibition, retest sample extract at 10-fold dilution. Cycling Protocol Name of Protocol: Serratia marcescens YV 1. 95 C for 180 sec 2. 45 cycles of: 95 C for 30 sec 55 C for 30 sec 72 C for 120 sec 3. Final extension 72 C for 600 sec 4. Melt curve from 60 C to 95 C, ramp rate 0.2 C/sec Duration of protocol: 2.75 h Expected Results Product is 409 bp long, with a Tm of 88.4-90.1ºC. Template-free controls in this assay have had a Tm of 62.3-64.4ºC. Non-specific products have had Tm values of 75.6-78.6ºC; some assays have yielded specific products of approximately 50 bp with Tm values of 78.0-81.1ºC. Limit of sensitivity with purified DNA is 10 pg genomic DNA per tube. Cells of Serratia marcescens appear to have thermostable nuclease enzymes that degrade the a79 amplicon, so put reaction tubes in ice immediately after PCR has run and gel the gel the same day, if needed. Otherwise, store PCR amplicons at -20 ºC. This has not been a problem with PCR runs using extracted DNA as the source of template. Reference Benny Bruton, pers. comm., 2003. Document Control: 2/13/16