Bisulfite Genomic DNA Sequencing
advertisement

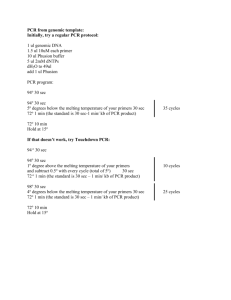
Bisulfite Genomic DNA Sequencing From Xiao, et al. 2003. “Imprinting of the MEA Polycomb gene is controlled by antagonism between MET1 methyltransferase and DME glycosylase.” And Jacobsen, et al. 2000. “Ectopic hypermethylation of flower-specific genes in Arabidopsis.” 1) Digest genomic DNA (0.3 – 0.7 g) in a 20 l reaction with restriction enzymes flanking the region of interest. 2) Add 30 g tRNA, and extract DNA once with phenol/chloroform. 3) Precipitate by adding NH4Oac (pH 7.0) to 3 M, plus 3 volumes EtOH. 4) Centrifuge. Wash pellet twice with 70% EtOH. Dry, and dissolve in 50 l TE. 5) Boil 2 min, and place on ice 1 min. 6) Treat with 2.2 l fresh 3 M NaOH, and incubate 15 min at 37 C. 7) Add 208 l bisulfite solution, and overlay with 3 drops mineral oil. Bisulfite solution: 40.5 g sodium bisulfite dissolved in 80 ml water, stirred slowly Adjust pH to 5.1 with freshly prepared 10 M NaOH Add 3.3 ml of 20 mM hydroquinone Adjust volume to 100 ml with water 8) Incubate in a thermal cycler for 30 cycles of 55C for 15 min and 95C for 30 sec. 9) For PCR, use 2 l bisulfite-treated DNA in a 50 l reaction. Use 400-600 nM primers, 0.5 l Taq DNA Polymerase, and 1 x dNTPs. PCR conditions are: 95C 5 min 5 cycles of 95C 15 sec, 60C 3 min, 72C 3 min 10 cycles of 95C 15 sec, 60C 1 min, 72C 2 min 30 cycles of 95C 15 sec, 50C 1 min 30 sec, 72C 2 min 72 C 5 min