Supplementary Material (doc 30K)
advertisement

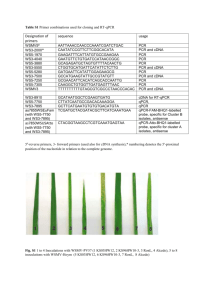
Supplementary Material 1) NPM1 mutation analysis For identifying NPM1 mutations, cDNA from reverse transcription was used. Exons 11 and 12 of NPM1 were amplified. The amplification mixture contained 2 µl of cDNA, 1X PCR Buffer, 2 mM MgCl2, 200 µM of each dNTP, 1.5 U of Taq Gold DNA polymerase (Applied Biosystems, Europe) cctggacaacatttatcaaacacggta-3’) and and 10 pmol of NPM1_1112 of the newly-designed NPM1_870 R1 F (5’(5’- tggttctcttcccaaagtggaa-3’) primers. 50 µl of the reaction mixture were subjected to the following thermocycler steps: 95°C for 7 min, 38 cycles of 30 s at 95°C, 30 s at 60°C, 30 s at 72°C and final extension of 7 min. All steps were carried out on a GeneAmp PCR System 2700 (Perkin Elmer, USA). DHPLC-based screening was done using a WAVE DNA fragment analysis system (Transgenomics Inc., San Jose, USA), through a 6.5 mm Ø DNAsep HT® Cartridge (Transgenomics). As first step, DHPLC was conducted in partially denaturating conditions optimized for exon-12, as recently described (Ammatuna E et al. Clin. Chem. 51:2165-2167, 2005). Amplicons showing an heteroduplex profile were purified (ExoSAP-IT®, Amersham Biosciences, Sweden) and sequenced directly in both strands (Big Dye Terminator Cycled Sequencing Kit, Perkin Elmer, USA) on an automated sequencer ABI Prism 310 (Perkin Elmer, USA). All homoduplex samples were retested under specific conditions for exons 9, 10 and 11. This amplification step was done using primers NPM1_1112 R and the newly-designed NPM1_658 F (5’- gaaaaagcgccagtgaagaaa-3’). DHPLC runs were performed at 54°C and 55°C, as suggested by the NavigatorTM software predicted melting curve (Ver 1.6.0) 2) Allele-specific oligonucleotide PCR (ASO-PCR) To confirm the identity of the mutation in exon-11, an allele-specific oligonucleotide polymerase chain reaction (ASO-PCR) was performed using the following reaction mixtures: 2 µl of patient cDNA and 2 µl of cDNA from two healthy controls were amplified by 10 pmol of the mutation-specific oligonucleotide (ASO) NPM1_882Fmut (5’caaagtggaagccaaattcaggcg-3’) and by 10 pmol of normal primer (NP) NPM1_881Fwt (5’ccaaagtggaagccaaattcatcaa-3’). 10 pmol of the common reverse primer NPM1_1112R was used for both reaction mixtures. Both reactions contained 1X PCR Buffer, 2 mM MgCl2, 200 µM of each dNTP, 1.5 U of Taq Gold DNA polymerase (Applied Biosystems). 50 µl of each reaction mixture were run on a GeneAmp PCR System 2700 (Perkin Elmer) at the following amplification steps: 95°C for 7 min, 35 cycles of 30 s at 95°C, 30 s at 65°C, 30 s at 72°C and final extension of 7 min. Mutated and wild-type amplification fragments were cut from agarose gel and purified with QIAquik® Gel Extraction Kit (Qiagen, USA) following the manufacturer’s instructions. Reverse strands of purified PCR products were directly sequenced as described above. 3) Cell cultures and transfection procedures NIH-3T3 murine fibroblasts were cultured in D-MEM supplemented with 10% Bovine Calf Serum (BCS), 1% glutamine and antibiotics. For transfection purposes, cells were seeded overnight on glass coverslips and transfected using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) following the manufacturer's instructions. After 24 hours’ incubation, cells were incubated with cycloheximide (Merck Biosciences Ltd, Nottingham, UK) 10 mg/ml for 30 minutes. Leptomycin B (Merck Biosciences), a specific CRm1 inhibitor (Fukuda M et al., Nature 390:308-311, 1997), was added at 20 ng/ml for 5 hours. 4) Immunofluorescence analysis For fluorescence studies, NIH 3T3 cells were rinsed in PBS containing 0.1% Triton X-100 and fixed in 4% paraformaldehyde pH 7.4 (10 minutes), washed, counterstained with propidium iodide, mounted on Mowiol and observed with a Zeiss (Carl Zeiss, Jena, Germany) LSM 510 confocal microscope, using a Plan Apochromat 100x/1.4 NA oil objective. Images were collected using 488-nm (for e-GFP) and 543-nm (for propidium iodide) laser lines for excitation. AOTF-controlled tuning of laser lines, pinhole diameters, and light collection configuration were optimized to obtain best signal-to-noise ratio and to avoid fluorescence crossover. LSM 510 software regulated the microscope; images were collected and transferred to an SGI Octane workstation (Silicon Graphics, Mountain View, CA) for further processing. Slices were reconstructed in 3D using the shadow technique or isosurface analysis, with Imaris software (Bitplane, Zurich, Switzerland), as previously described (Quentmeier H et al., Leukemia 19:1760-1767, 2005). The cells were cut electronically to analyse localization of NPM mutants. 5) Immunohistochemical detection of NPM Nucleophosmin was detected on bone marrow paraffin sections using monoclonal antibodies directed against fixative-resistant epitopes of NPM (clones 322 and 376 produced in B. Falini’s laboratory) and C23/nucleolin (Santa Cruz, Biotechnology, Heidelberg, Germany), as previously described (Falini B et al., N Engl J Med 352:254-266, 2005). Paraffin sections were also stained with monoclonal antibodies directed against glycophorin, myeloperoxidase, and CD68 (all purchased from Dako Cytomation, Glostrup, Denmark). In all instances, antibody-antigen binding was revealed using the immunoalkaline phosphatase APAAP technique (Cordell JL et al., J Histochem Cytochem 32:219229, 1984).