cDNA Reaction
advertisement

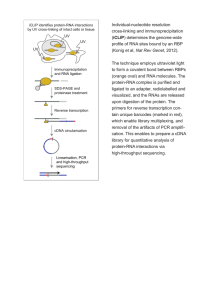
cDNA Reaction Materials and reagents: iScript Reaction Mix (xxxxxx) RNase away (Fisher) or equivalent (RNA Zap may be found around the lab – this is not preferred, as it has a tendency to foam) 200 ul PCR tubes (USA Scientific) Aerosol resistant filter tips, 20 ul, 200 ul and 1000 ul (various suppliers) – use only newly opened, pre-sterilized boxes – do not use autoclaved tips RNase-free water (LifeTechnologies or Ambion) Reaction set up: Component Volume per reaction 5x iScript Reaction Mix iScript Reverse Transcriptase RNA template (100fg to 1ug total RNA) Nuclease-free water 4 ul Total volume 20 ul 1 ul x ul (15-x) ul Reaction cycle: Incubate the complete reaction mix (use a thermal cycler): 5 minutes at 25°C 30 minutes at 42°C 5 minutes at 85°C Hold at 4°C until reactions can be removed. In addition to the actual samples, standard control reactions are: iScript Reaction Mix + Reverse Transcriptase + H2O to check for kit contamination iScript Reaction Mix + RNA template + H2O to check for DNA contamination of sample For quality control and to check for DNA contamination, run a standard PCR reaction on the cDNA samples, using a primer set known to work, and preferably one to a gene that would be expressed in the sample conditions. The complete cDNA reactions should have amplification, while the controls should not. Note: cDNA does not react well to multiple freeze-thaw cycles, so minimize the number of times you use a single sample.