TPJ_3608_sm_AppendixS3
advertisement

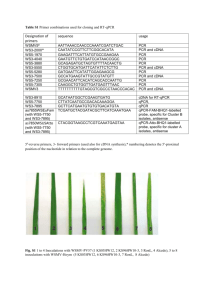
Appendix S3: Experimental procedures Materials Wild-type and T-DNA insertion lines of Arabidopsis were obtained from the European Arabidopsis Stock Centre (Scholl et al., 2000), except for the u19-2 homozygous line (WiscDsLox430F05) which was kindly provided by P. Goldsbrough, Purdue University. DNA isolation and PCR reactions DNA was isolated from WT and T-DNA insertion lines using CTAB method (Murray and Thompson 1980), or a buffer containing SDS (200 mM Tris-HCl pH 7.5, 250 mM NaCl, 25 mM EDTA, 0.5% SDS), followed by precipitation in 2propanol and 70% ethanol. Amplification conditions were: 4 min at 94°C, followed by 30 to 35 cycles of 20 sec at 94°C, 30 sec at 60°C, 1 minute at 72°C, and an extension of 5 min at 72°C after the final cycle. Amplification was carried out in a 50 μl volume consisting of final concentration of 1X PCR buffer (supplied with the Promega Taq polymerase, Catalog # M8305), 2 mm MgCl2, 0.2 mM deoxynucleotide (dNTP) mixture (Bioline), 0.2 μM each sense and antisense primer, 2.5 units Taq polymerase (Promega), and 50-100 ng template DNA. Reaction products were separated by electrophoresis on a 1% agarose gel in 1X TAE buffer (Sambrook et al., 1989). Fermentas GeneRuler 1 kb DNA ladder (SM0311) was used as size marker in agarose gels. Primers used in PCR assays are given in supplementary table 1. 1 Measurement of gene expression Total RNA was prepared from 100 mg two week-old seedling tissue using RNeasy plant RNA minipreps (Qiagen, Dorking, UK) or from whole plants using Tri reagent (Sigma). For gel blot analysis, 10 µg per sample was electrophoresed through 1.0% agarose (Life Technologies, Paisley, UK) formaldehyde gels (Sambrook et al., 1989). RNA was transferred to nylon membranes (Roche, Mannheim, Germany) by capillary action. Blots were pre hybridised and hybridised in 50% formamide as described previously (Knight et al., 1999) at 42C. Blots were washed twice in each of the following successively: 2X SSC (1X SSC is 0.15 M sodium chloride, 0.15 M sodium citrate, pH7), 0.1% SDS followed by 1X SSC, 0.1% SDS and finally 0.1X SSC, 0.1 % SDS at 42C. Probes were prepared by PCR or digestion of ESTs and labelled with 32P-CTP by the random priming method using DNA “Ready-to-go” labelling beads (GE Healthcare, Little Chalfont, UK). For semi-quantitative RT-PCR assays cDNA synthesis was performed on 5 g total RNA using a cDNA synthesis kit (Fermentas, St. Leon-Rot, Germany, catalog number K1632) with 5 μM genespecific antisense or sense primer and 5 μM anchored oligo dT primer or genespecific sense or antisense primers and 10 units of MMLV reverse transcriptase. cDNA was diluted 10-20 fold with water and 2-5 μl was used in semi-quantitative PCR assays. PCR conditions were the same as for genomic DNA PCR but the cycle numbers varied. For realtime-PCR assays, cDNA was prepared from 1 µg RNA using random hexamer primers and Superscript II reverse transcriptase (Invitrogen) according to the manufacturer’s protocol. cDNA was treated with 2 RNase H and RNase A after reverse transcription. Quantitative real-time PCR was carried out using a SYBR green mastermix (Applied Biosystems) and an ABI Prism 7000 Sequence Detection System (Applied Biosystems). TPC1 and UBQ10 (At4g05320) expression levels were determined by absolute quantification against a standard curve derived from a cDNA dilution series. TPC1 expression levels were normalized using UBQ10. Realtime-PCR conditions for all primer combinations were: 95°C for 10 min; 40 cycles of (95°C for 15 sec, 60°C for 1 min), followed by a melting curve protocol to determine the specificity of primers. Primers used in real-time PCR are given in supplementary table 1. Proteomic analysis of glutathione transferases For each line, approx. 4 g of root culture tissue (DeRidder et al., 2002) was harvested and glutathione transferases were purified using glutathione affinity chromatography as described (DeRidder et al., 2002). The purified samples were analysed by 2D-PAGE as described (DeRidder et al., 2002) except that the entire sample was used and 12% acrylamide gels were used for 2nd dimension separation. Gels were stained with Coomassie Brilliant Blue and proteins in the stained spots were identified by peptide mass fingerprinting (Loutre et al., 2003). 3 References for the Appendix S3 DeRidder, B.P., Dixon, D.P., Beussman, D.J., Edwards, R., Goldsbrough, P.B. (2002) Induction of glutathione S-transferases in Arabidopsis by herbicide safeners. Plant Physiol. 130, 1497-505. Knight, H., Veale, E., Warren, G.J. and Knight, M.R. (1999) The sfr6 mutation in Arabidopsis suppresses low-temperature induction of genes dependent on the CRT/DRE sequence motif. Plant Cell, 11, 875–886. Loutre, C., Dixon, D.P., Brazier, M., Slater, M., Cole, D.J., Edwards, R. (2003). Isolation of a glucosyltransferase from Arabidopsis thaliana active in the metabolism of the persistent pollutant 3,4-dichloroaniline. Plant J. 34, 485-93. Murray, M.G. and Thompson, W.F. (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8, 4321-4325. Sambrook, J., Fritsch, E.F. and Maniatis, T. (1989) Molecular Cloning. A Laboratory Manual. New York: Cold Spring Harbour. Scholl, R.L., May, S.T., Ware, D.H. (2000) Seed and molecular resources for Arabidopsis. Plant Physiol. 124, 1477-1480. 4