A highly sensitive dHPLC technique reveals PIK3CA
advertisement

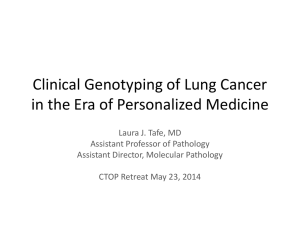
Supplemental Methods, Results and Figure Legend: Supplemental Methods: SSCP, dHPLC Analysis and Sequencing of PIK3CA Amplifications for dHPLC analysis of PIK3CA, exon 9 were carried out with primers X9FF and X9RR, and primers X20F2 and X20R2 for exon 20. For PIK3CA sequence analysis, exon 9 was amplified with primers X9F and X9RC, and exon 20 with primers X20F1 and X20R. PCRs were performed in a 50-l volume containing 50 ng genomic DNA, 2 mM MgSO4, 140 M dNTPs, 0.3 M of each primer, and 2.5 U Platinum Hi Fidelity DNA Taq polymerase (Invitrogen). The initial denaturing step at 94C for 2 min was followed by 35 cycles consisting of 30 sec at 94C, 30 sec at 55C, and 45 sec at 68C. The final extension step at 72C was 2 min. PCR was performed in a GeneAmp 2400 PCR System (Applied Biosystems). PCR products were purified on a Qiagen QiaQuick column and sequenced at the Moores-UCSD Cancer Center shared sequencing resource with internal primers X9S and X9SF and X20F and X20S for exons 9 and 20, respectively. For SSCP analysis of the region of PIK3CA exon 20 that most frequently harbors mutations, DNA was amplified with primers X20F2 and X20R2 using 50 ng of genomic DNA in a 50 l reaction volume containing: 1X Qiagen PCR buffer, 2.0 mM MgCl2, 50 M 4dNTP, 50 Ci α32P-dATP, 1 unit Taq DNA polymerase (Qiagen) and 10 pmol each sense and antisense primer. Following an initial denaturation at 95oC for 3 min., amplification proceeded for 30 sec. each at 95oC, 58oC and 72oC for 30 cycles. The amplification products are diluted 50-fold in 0.1% sodium dodecyl sulfate (SDS), 10 mM EDTA, diluted a further 2-fold in 2X loading buffer (95% formamide, 20 mM EDTA and 20 mM NaOH), heat denatured, cooled and resolved on a 0.5x MDE gel containing 5% glycerol in 0.6X TBE (54 mM Tris-borate, 1.2 mM EDTA) and resolved at 8W for 18h at room temperature. Denaturing high-performance liquid chromatography (dHPLC) was performed using the WAVE DNA Fragment Analysis System (Model 3500 HT; Transgenomic), controlled by Navigator software. To enhance heteroduplex formation, PCR products were denatured at 95C for 5 minutes, followed by gradual reannealing to 25C at -0.1C/sec. Initially, for each PCR product, the melting behavior of the wild-type sequence was visualized using the WAVEMAKER software (Transgenomic) algorithm. The elution for each PCR product was performed at a temperature corresponding to 80-90% -helical fraction for each melting domain. After a few test trials with the positive control samples for each axon, the optimal temperature for mutation detection for 258-bp exon 9 was determined to be 59C, and 58C for 420-bp exon 20. Sample volumes of 10 l (optimal quantity of the loaded PCR product corresponded to a peak of A260 ~4-12 mV) were loaded on a DNASep (Transgenomic) cartridge and eluted within 4.5 min at a flow rate of 0.9 ml/min using a linear acetonitrile gradient 48.3 – 62.3% buffer B (0.1 M triethylammonium acetate [TEAA]; 25% acetonitrile) for exon 9 PCR products and 50.8 – 68.4% buffer B for exon 20 PCR products. Eluted DNA fragment was detected by the system’s ultraviolet detector and observed online at 260 nm. Regeneration of the column was achieved by washing with 100% buffer D (75% acetonitrile) for 30 sec followed by an equilibration time of 2 min. The ability and sensitivity of this protocol is illustrated in the Supplemental Figure. Primers: (5’ 3’) X9F GATTGGTTCTTTCCTGTCTCTG X9RC CCACAAATATCAATTTACAACCATTG X9S TTGCTTTTTCTGTAAATCATCTGTG X9SF AGTTTAAAAATCATGTAAATTCTGCTT X9FF CCAGAGGGGAAAAATATGACA X9RR TGCTGAGATCAGCCAAATTC X20F1 TCTAGTGGGGTAAAGGGAATCA X20R GGGGATTTTTGTTTTGTTTTG X20F2 GACCTGAAGGTATTAACATCATTTGC X20R2 ATTCCTATGCAATCGGTCTTG X20F TTGCATACATTCGAAAGACC X20S TTTGTTTTGTTTTGTTTTTT Supplemental Results No leukemia cell lines were found to have PIK3CA mutations. In primary T-ALL, a heterozygous conserved mutation at codon 546 in exon 9 (CAG (Gln) (C/A)AG (Gln/Lys)) and a heterozygous silent mutation at codon 1025 in exon 20 (ACC (Thr) AC(C/T) (Thr/Thr)) were identified. Several sequence changes outside of the coding or splice region of exon 9 were also identified in the 46 patient samples sequenced. A T T/G located 105 bases 3’ of the exon 9 coding sequence was observed in 3 T-ALL cell lines and 7 primary ALL. A CC/T located 85bp downstream of exon 9 was observed in one patient and a CC/T located 55 bp upstream of exon 9 was observed in 4 patients. Their presence in non-coding regions suggest they are polymorphisms and their characterization was not pursued further. Supplemental Figure Legend dHPLC detection of PIK3CA mutations in PIK3CA exons 9 and 20. DNA obtained from the MCF7 breast cancer cell line was used as a positive control for mutations to PIK3CA exon 9, and DNA from the LS174 colon cancer cell line was used as a positive control for mutations to PIK3CA exon 20. dHPLC analysis of PIK3CA exon 9 from MCF7 DNA (panel A) and exon 20 from LS174 DNA (panel B) show distinct elution profiles versus DNA from normal cells, suggesting the presence of mutations. The exon 20 mutation in LS174 is also easily detectable via SSCP (panel C) (lane 1) versus the HL60 (lane 2) and primary leukemia samples (lanes 3-5) with a wild-type PIK3CA exon 20. Mutations were confirmed by direct sequence analysis (PIK3CA exon 9 from MCF7 (G G/A mutation at bp 1633, codon 545 (panel D) and PIK3CA exon 20 from LS174 (A A/G mutation at bp 3140, codon 1047 (panel E). Sensitivity of the dHPLC assay was determined by the sequential dilution of MCF7 DNA, which carries a PIK3CA exon 9 mutation, with DNA from a normal individual (panel F). This assay could detect a heterozygous mutation in a DNA sample diluted 20-fold with normal DNA, representing the ability to detect a heterozygous mutation present in a population containing as few as 5% tumor cells, or a homozygous mutation in a sample population containing as few as 2.5% tumor cells.