emi412036-sup-0004-text1

1 Supplementary text 1: Detailed experimental procedures
2 Insects
3 Experimental M. separata were from a laboratory colony originated with insects
4 collected in 2009 on corn near Xinxiang City (35°18′N, 113°52′E), Henan province,
5 China. Ten larvae/glass jar (9x13 cm) were mass reared on maize seedlings (30-50 cm
6 high) at 23±1°C, 60-80%RH and 14:10L/D photoperiod (Luo et al ., 1995). Different
7 instar larvae were randomly selected for experiments. Larvae were chilled on ice for 5
8 min, and their midguts isolated and washed with physiological saline (NaCl, 0.15
9 mol/L). The samples were immediately frozen in liquid nitrogen, and then stored at
10 -70°C until use.
11 Cloning the full-length MsCAD1 cDNA
12 RT-PCR and RACE were used to clone full-length MsCAD1 cDNA. Total RNA was
13 isolated using Trizol reagent (Invitrogen) according to the manufacturer’s instructions.
14 First-strand cDNA was synthesized using the Quantscript RT kit (TransGen Biotech,
15
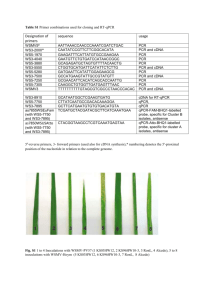
Beijing, China) following the manufacturer’s protocol. The resulting cDNA was used
16 as PCR template. Two pairs of degenerate primers (listed in Table S1; CADF1 and
17 CADR1, CADF2 and CADR2) were designed based on a conserved region in CADs
18 of other insects. MsCAD1 gene fragments were amplified with Ex-Taq (TAKARA,
19 Dalian, China), programmed at 30 cycles at 94°C for 30s, 51°C for 45s, and 72°C for
20 1 or 2 min 20s, followed by a final extension at 72°C for 10 min. Purified PCR
21 products were inserted into the pEASY-T3 Cloning Vector (TransGen Biotech,
22 Beijing, China) and sequenced by a commercial laboratory (Beijing Aoke, Beijing,
23 China).
24 Additional primers were designed on the basis of the PCR product sequence
25 (relative primer positions shown in Fig. S2). 3′- and 5′-RACE PCR was carried out
26 under the conditions just described using LA-Taq (TAKARA, Dalian, China). The
27 amplified products were inserted into the pEASY-T3 Cloning Vector (TransGen
28 Biotech, Beijing, China) and sequenced (Beijing Aoke). For quality assurance relative
29 to sequencing errors, specific primer pairs (QCF and QCR) were used to amplify the
30 predicted gene including the entire open reading frame, programmed at 30 cycles at
31
98°C for 10s, 68°C for 320s, followed by a final extension at 72°C for 10 min.
32 Sequence alignment and identity analysis
33 Full-length MsCAD1 cDNA was used to interrogate GenBank by BLAST software
34 (http://www.ncbi.nlm.gov/blast/). Sequence alignment and identity analysis were
35 carried out using the DNAMAN (version 6.0) software, and ORFs were analyzed at
36 http://www.ncbi.nlm.nih.gov/gorf/orfig.cgi. Molecular weights and isoelectric points
37 of the predicated proteins were calculated by Protparam software
38 (http://expasy.org/tools/protparam.html). Signal peptide and transmembrane domain
39 prediction were done using the SignalP 3.0 Server
40 (http://www.cbs.dtu.dk/services/SignalP/) and the TMHMM Server v. 2.0
41 (http://www.cbs.dtu.dk/services/TMHMM/), respectively. A motif scan of the protein
42 sequence was conducted using Hits software (http://hits.isb-sib.ch/cgi-bin/PFSCAN?).
43 An unweighted pair group method with arithmetic mean (UPGMA) phylogenetic tree
44 was reconstructed using MEGA software under the Poisson correction model.
45 Bootstrap values are expressed as percentages of 500 replications.
46 Real-time fluorescence quantitative PCR
47 Total RNA was extracted using Trizol (Invitrogen) and quality and quantity were
48 assessed by agarose gel electrophoresis and spectrophotometry (NanoDrop 2.5.1).
49 One microgram of total RNA from each sample was reverse transcribed with the
50
PrimeScript® RT reagent Kit with gDNA Eraser (Perfect Real Time, TAKARA). Two
51 pairs of primers (YGF and YGR, ACF and ACR) were designed based on the highly
52 conserved region of the MsCAD1 cDNAs and the M. separata
β
actin gene using
53 Ex-Taq polymerase (TAKARA) and the corresponding primers (Table S1)
54 programmed at 30 cycles at 94°C for 30s, 60°C for 30s, and 72°C for 30s, followed
55 by a final extension at 72°C for 10 min. The PCR amplification products were
56 analyzed, cloned and sequenced as just described. RT-qPCR for MsCAD1 and
β
actin
57 genes were carried out on an iCycler iQ (Bio-Rad, USA) using SYBR Premix Ex-Taq
58
(TAKARA) under the following conditions: 95°C for 30s, followed by 40 cycles at
59 95°C for 5s, and 60°C for 45s. Specificity of the SYBR Green RT-PCR was assessed
60 by generating a melting curve using the dissociation curves obtained from the PCR
61 products and the derivatives (−dF/dT) of fluorescence values plotted at 0.5°C
62 intervals from 60°C to 95°C.
63 The qPCR standard curves constructed in this study were used to quantify
64 expression data of MsCAD1.
The recombinant plasmid concentration was determined
65 by spectrophotometry (NanoDrop 2.5.1), then serially diluted 10-fold. Three technical
66 replicates were used to produce the standard curve, and three independent biological
67 replicates were conducted for each sample. A no-template control was included in all
68 SYBR Green quantification RT-PCR assays. The
β
actin reference gene, expressed at
69 the same level in several independent runs, was used as an endogenous reference gene.
70 The average concentration across sample replicates was obtained from the standard
71 curves and the relative expression ratio of the target gene was calculated according to
72 protocols established by the manufacturer (ABI Prism 7700 Sequence Detection
73 System). The mathematical models used for relative quantification of the target gene
74 were described by Rasmussen (2001) and Pfaffl (2001), and the PCR efficiencies
75 were between 95% and 105%, the effective amplification efficiency scope. All the
76 numerical data obtained from the quantitative tests are presented as mean ± SEM.
77 Statistical analyses were conducted using SAS V. 8.0 software, applying one-way
78 ANOVA and Tukey’s HSD test. Significance was set at P <0.05 and all differences
79 mentioned are statistically significant.
80 RNA interference
81 Three pairs of MsCAD1 -specific primers (ds1F and ds1R, ds2F and ds2R, ds3F and
82 ds3R; TableS1), all containing the T7 promoter sequence (5′
83
TAATACGACTCACTATAGGG 3′), were designed to synthesize three
84 double-stranded RNA (dsRNA) sequences. MsCAD1 cDNAs were used as template in
85 PCR reactions to amplify 414, 423 and 500-bp fragments, denoted ds1, ds2 and ds3 in
86 the results section. The expected size of these PCR products was verified by agarose
87 gel and then used for in vitro transcription of the dsRNA using the MEGAscript® T7
88 Kit (Ambion, USA) following the manufacturer’s protocol. After purification, the
89 dsRNA was diluted in water and quantified using spectrophotometry as described
90 above.
91 We used 6-well plates as feeding chambers and 1.5-ml centrifugal tube caps as
92 feeding containers. Each cap was filled with artificial diet and covered evenly with a
93
10 μl dsRNA water solution. The dsRNA concentrations were set at two doses, a high
94
(0.25 μg/μl) and a low dose (0.05 μg/μl). First to 3 rd
-instar larvae were selected and
95 fasted 6 hours before initiation of the assays. Ten individuals were transferred into an
96 individual well. Sets of 60 individuals comprised one replicate and three replicates
97 were conducted. The artificial diet was replaced daily to minimize the possibility of
98 dsRNA degradation in test samples.
99 Semi-quantitative RT-PCR was performed to observe expression of MsCAD1 at
100 selected time points after dsRNA treatments. Total RNA was isolated after 0, 24, 48,
101 72 and 96 hours of dsRNA consumption, using about 20 larvae for each time point,
102 and cDNA was synthesized. Semi-quantitative RT-PCR reactions were performed
103 using the primers just described, programmed at 25 cycles at 98°C for 10s, 52 ℃ for
104
30s, 72°C 1 min; followed by a final extension at 72°C for 10 min; for the β actin
105 internal control, the PCR program was 25 cycles at 98°C for 10s, 60 ℃ for 45s,
106 followed by a final extension at 72°C for 10 min. The PCR products were analyzed on
107 agarose gel electrophoresis and stained with ethidium bromide. Total RNA (1μg each)
108 was reversed transcribed to cDNA using an oligo dT
18
primer from the midgut. One
l
109 cDNA was used as a template for PCR reactions using MsCAD1 specific primers (25
110 cycles) or β actin specific primers (25 cycles). The loading volume from all the
111 treatments for agarose gel electrophoresis was 10
l.
112 Insect bioassays
113 We used 3 rd
instar larvae for all bioassays. Purified (> 98%) activated B. thuringiensis
114 Cry1Ab toxin was obtained from Case Western Reserve University (Cleveland, OH).
115 Control larvae were fed routine diets covered evenly with 10 μl water (no dsRNA),
116 and experimental larvae were fed routine diets with the same volume of 0.25 μg/μl
117 dsRNA water solution. After 18 h exposure to the diets, control and experimental
118 larvae were fasted 2 h and then placed on routine diets (controls) and routine diets
119 amended with 45 μg Cry1Ab toxin/g diet (experimentals; the lethal concentration for
120 50% (LC
50
) of 4th instar larvae; Jiang et al.
, 2010) using new 6-well plates. The plates
121 were held as described above. Larval mortality and weight were determined after 7
122 days and the duration of 3rd instar was recorded. Each treatment was conducted in
123 three independent replicates with 60 larvae in each replicate. Mortalities in each
124 treatment were corrected relative to their corresponding mortality on the routine diet
125 following the Abbott formula (Abbott, 1925); data is presented as mean ± SEM. The
126 mortality proportions were arcsin transformed. A one-way ANOVA was performed to
127 determine treatment differences. Significant in differences among multiple means
128 were determined by Tukey’s HSD ( P <0.05). All statistical procedures were performed
129 with SPSS 11.0 software.
130 Expression and purification of recombinant MsCAD1
131 Three pairs of specific primers (CR9F/CR12R, CR9F/CR10R and CR11F/CR12R)
132 were designed to amplify MsCAD1 cDNA fragments of different lengths. To facilitate
133 cloning, two restriction enzyme sites ( Nde Ⅰ and Eco R Ⅰ ) were introduced. PCR
134 reactions were programmed at 30 cycles at 95°C for 30s, 54°C for 30s, 60°C for 60s
135 or 90s; followed by a final extension at 72°C for 10 min, using FastPfu DNA
136 Polymerase (TransGen Biotech, Beijing, China). The PCR products were purified and
137 subcloned into the expression vector pET30a. Positive clones were identified by
138 restriction analysis and sequencing. The three fragments with their restriction sites
139 were cloned into a pET30a expression vector, which was digested with the same
140 restriction enzymes. The recombinant plasmids were transformed into E. coli BL21
141 (DE3) competent cells. A verified single colony was grown overnight in 5-mL LB
142 medium amended with kanamycin (100 μg/mL). The culture was added to 500ml
143 fresh LB medium and grown at 37°C until OD600 reached 0.6-1.0, then 0.3mM
144 isopropyl -D-1thiogalactopyranoside was added to the culture to induce expression
145 of the target protein. The induced cells were incubated at 37°C for 4 h and harvested
146 by centrifugation. The pellets were resuspended in lysis buffer (10 mM imidazole,
147 300 mM NaCl and 50 mM NaH
2
PO
4
, PH 8.0) at 2 to 5 ml/g wet weight and sonicated
148 on ice after adding lysozyme. The supernatant was mixed with Ni
2+
-NTA resin,
149 washed with wash buffer (pH 8.0) containing 50 mM NaH
2
PO
4
, 300 mM NaCl and 20
150 mM imidazole to remove impurities. Finally, the recombinant His
6
-tagged proteins
151 were eluted with elution buffer (250 mM imidazole, 300 mM NaCl and 50 mM
152 NaH
2
PO
4
, pH 8.0). The purified proteins were analyzed by SDS-PAGE.
153 Western blot analysis
154 Purified proteins (with the His-tag) were separated by 12% SDS-PAGE and
155 electroblotted onto a nitrocellulose membrance (Protran Dassel, Germany) for 2 h at
156 80 V. The membrane was blocked with 5% nonfat powdered milk in Tris-buffered
157 saline containing 0.05% Tween 20 (TBS-T), and incubated with activated Cry1Ab
158 toxin at 37°C for 2 h. After washing the unbound Cry1Ab three times in PBS and
159 once in TBS-T, 10 min each, the membrane was incubated with rabbit anti-Cry1Ab
160 antiserum (source) diluted 1:2,000 in blocking buffer overnight at 4°C. The
161 membrane was washed three times in PBS and once in TBS-T, 10 min each, and then
162 incubated with horseradish peroxidase conjugated goat anti-rabbit IgG secondary
163 antibody (Biosynthesis Biotech. Co., Ltd. Beijing) diluted 1:5000 in blocking buffer
164 for 1 h at 37°C. The membrane was washed three times with TBS-T and once in PBS,
165 10 min each, and then developed in DAB substrate buffer for 10 min. Western
166 blotting was replicated three times.
167
References
168
169
Abbott, W. S. (1925) A method for computing the effectiveness of an insecticide. J
Econ Entomol 18: 265-267.
170
171
172
173
Jiang, S.J., Luo, L.Z., Hu, Y., and Zhang, L. (2010) Effects of Cry1Ac protein on growth and development, reproduction and flight potential of the oriental armyworm,
Mythimna separata (Walker) (Lepidoptera: Noctuidae). Acta Entomol Sin 53 :
1360-1366.
174
175
176
Luo, L.Z., Xu, H.Z., and Li, G.B. (1995) Effects of rearing density on the food consumption and utilization of larval oriental armyworm Mythimna separata (Walker).
Acta Entomol Sin 38 : 428-435.
177
178
Pfaffl, M.W. (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29 : 2002-2007.
179
180
181
182
Rasmussen, R. (2001) Quantification on the light cycler. In: Meuer S, Wittwer C,
Nakagawara K (eds) Rapid cycle real-time PCR, methods and applications. Springer,
Heidelberg 1 : 21-34.