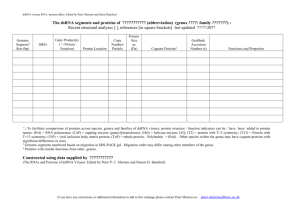
The dsRNA genome segments and proteins of
advertisement

dsRNA viruses RNA / protein tables: Edited by Peter Mertens and Denis Bamford
The dsRNA genome segments and proteins of Pseudomonas phage Phi 6
(genus Cystovirus : family Cystoviridae)
last updated 11/09/2002
dsRNA
(Size, bp)
[5,8,20,23]
Open reading
frames
(ORFs)
Proteins
(': protein
structure/
function)
Size 'aa'
(kDa )
[5,8,20,23]
protein copy
number per
particle
location
Function
Self assembles with proteins P2, P4 and P7 into procapsid particles
[9, 29]. Corresponds to the T=2 particle in BTV [11]. Controls
procapsid size and organization. Binds minor proteins and RNA.
Conserved in dsRNA viruses.
Semi-conservative RNA dependent RNA polymerase [34, 35].
Isolated protein acts on ss and dsRNA templates [18, 19]. Threedimensional structure homologous to HIV reverse transcriptase and
hepatitis C polymerase [3]. Binding sites for nucleic acid, NTPs,
Mn and Mg ions known [3].
Binds to the receptor [25] , the pilus IV of Pseudomonas syringae
[30, 31]
[5,8,20,23]
L
(6,374)
Gene 1
P1
(T=2)
769
(84.986)
120
[2. 6]
inner particle shell
L
Gene 2
P2 (pol)
[3, 18, 19]
664
(74.791)
12
[6]
inner particle
M
(4,063)
Gene 3
P3 (spike)
[17]
648
(69.178)
L
Gene 4
P4 (NTPase)
[7, 10, 16]
331
(35.032)
S
(2.948)
Gene 5
P5 (mur)
[4, 12, 21]
219
(24.018)
between the membrane and NC
[12]
M
Gene 6
L
Gene 7
S
Gene 8
P6 (fus)
[1]
P7 (assembly
factor) [29]
P8 (T=13)
[2]
167
(17.229)
160
(17.169)
148
(15.873)
Membrane
[32]
inner particle
[15]
second protein layer
[2]
S
Gene 9
89
(9.482)
M
Gene 10
P9 (env)
[13, 32, 33]
P10 (lys)
[14]
42
(4.272)
72
[7]
60
[15]
600
[2, 29]
extends from virion surface [17]
and is attached to the integral
membrane protein P6 [25]
inner particle at 5 fold axes
[7]
Membrane
[32]
Membrane
[32]
Hexameric NTPase, symmetry mismatched position [7, 10, 16].
Powers the packaging of ssRNA genomic precursors and is
involved in transcription [27]. Nucleates the assembly of the
procapsid with P1 [29]
Muralytic enzyme exposed to host peptidoglycan layer after
membrane fusion during viral entry. Also used to lyse the host cell
late in infection to release the virus [4, 12, 21].
Integral membrane protein that has membrane fusion activity and
binds the receptor -binding protein, P3 [1, 25, 32].
Dimeric, stabilizes the procapsid [15]. Increases the assembly rate
of the procapsid in vitro [29]
A trimer [29] that forms the T=13 nucleocapsid shell around the
dsRNA-containing inner particle (core) [2]. During entry P8 drives
the penetration of the nucleocapsid into the cytoplasm [26, 28].
Major membrane protein, essential for membrane formation [13, 32,
33]
Needed for cell lysis [14]
dsRNA viruses RNA / protein tables: Edited by Peter Mertens and Denis Bamford
S
Gene 12
M
Gene 13
P12
(assembly)
[13. 22, 25]
P13
L
Gene 14
P14
195
(20.293)
72
(7.649)
61
(6.810)
0
0
Non-structural
[32]
Assembly factor active in viral membrane morphogenesis [13, 22,
25].
Membrane
[8]
Non- structural
[5]
Minor, nonessential membrane protein [8, 24]
Nonessential [5].
': ': To facilitate comparisons of proteins across species, genera and families, protein structure / function indicators have been added to
protein names, eg (pol) = RNA polymerase; (T=2) = capsid protein organized with icosahedral T=2 symmetry; (T=13) = capsid protein
organized with icosahedral T=13 symmetry.
Constructed by Sarah Butcher and Dennis Bamford.
Available as a webpage on: http://www.iah.bbsrc.ac.uk/dsRNA_virus_proteins/Cystovirus.htm
If you have any queries regarding this page please contact Peter Mertens on peter.mertens@bbsrc.ac.uk.
References.
1. Bamford, D.H., Romantschuk, M. and Somerharju, P.J. (1987) Membrane fusion in prokaryotes: bacteriophage phi6 membrane fuses with the
Pseudomonas syringae outer membrane. EMBO J., 6, 1467-1473.
2. Butcher, S.J., Dokland, T., Ojala, P.M., Bamford, D.H. and Fuller, S.D. (1997) Intermediates in the assembly pathway of the double-stranded
RNA virus f6. EMBO J., 16, 4477-4487.
3. Butcher, S.J., Grimes, J.M., Makeyev, E.V., Bamford, D.H. and Stuart, D.I. (2001) A mechanism for initiating RNA-dependent RNA
polymerization. Nature, 410, 235-40.
4. Caldentey, J. and Bamford, D.H. (1992) The lytic enzyme of the Pseudomonas phage phi 6. Purification and biochemical characterization.
Biochim Biophys Acta, 1159, 44-50.
5. Casini, G. and Revel, H.R. (1994) A new small low-abundant nonstructural protein encoded by the L segment of the dsRNA bacteriophage phi
6. Virology, 203, 221-8.
6. Day, L.A. and Mindich, L. (1980) The molecular weight of bacteriophage phi6 and its nucleocapsid. Virology, 103, 376-385.
dsRNA viruses RNA / protein tables: Edited by Peter Mertens and Denis Bamford
7. de Haas, F., Paatero, A.O., Mindich, L., Bamford, D.H. and Fuller, S.D. (1999) A symmetry mismatch at the site of RNA packaging in the
polymerase complex of dsRNA bacteriophage phi6. J Mol Biol, 294, 357-72.
8. Gottlieb, P., Metzger, S., Romantschuk, M., Carton, J., Strassman, J., Bamford, D.H., Kalkkinen, N. and Mindich, L. (1988a) Nucleotide
sequence of the middle dsRNA segment of bacteriophage phi 6: placement of the genes of membrane-associated proteins. Virology, 163, 18390.
9. Gottlieb, P., Strassman, J., Bamford, D. and Mindich, L. (1988b) Production of a polyhedral particle in Escherichia coli from a cDNA copy of
the large genome segment of Phi 6. J. Virol., 62, 181-187.
10. Gottlieb, P., Strassman, J. and Mindich, L. (1992) Protein P4 of the bacteriophage phi 6 procapsid has a nucleoside triphosphate-binding site
with associated nucleoside triphosphate phosphohydrolase activity. J. Virol., 66, 6220-2.
11. Grimes, J.M., Burroughs, J.N., Gouet, P., Diprose, J.M., Malby, R., Zientara, S., Mertens, P.P. and Stuart, D.I. (1998) The atomic structure of
the bluetongue virus core. Nature, 395, 470-8.
12. Hantula, J. and Bamford, D.H. (1988) Chemical crosslinking of bacteriophage ø6 nucleocapsid proteins. Virology, 165, 482-488.
13. Johnson, M.D. and Mindich, L. (1994a) Plasmid-directed assembly of the lipid-containing membrane of bacteriophage phi 6. J Bacteriol, 176,
4124-32.
14. Johnson, M.D. (3rd) and Mindich, L. (1994b) Isolation and characterization of nonsense mutations in gene 10 of bacteriophage phi 6. J. Virol.,
68, 2331-8.
15. Juuti, J.T. and Bamford, D.H. (1997) Protein P7 of phage phi6 RNA polymerase complex, acquiring of RNA packaging activity by in vitro
assembly of the purified protein onto deficient particles. J Mol Biol, 266, 891-900.
16. Juuti, J.T., Bamford, D.H., Tuma, R. and Thomas, G.J., Jr. (1998) Structure and NTPase activity of the RNA-translocating protein (P4) of
bacteriophage phi 6. J Mol Biol, 279, 347-59.
17. Kenney, J.M., Hantula, J., Fuller, S.D., Mindich, L., Ojala, P.M. and Bamford, D.H. (1992) Bacteriophage phi 6 envelope elucidated by
chemical cross-linking, immunodetection, and cryoelectron microscopy. Virology, 190, 635-644.
18. Makeyev, E.V. and Bamford, D.H. (2000a) The polymerase subunit of a dsRNA virus plays a central role in the regulation of viral RNA
metabolism. EMBO J., 19, 6275-6284.
dsRNA viruses RNA / protein tables: Edited by Peter Mertens and Denis Bamford
19. Makeyev, E.V. and Bamford, D.H. (2000b) Replicase activity of purified recombinant protein P2 of double- stranded RNA bacteriophage phi6.
EMBO J, 19, 124-133.
20. McGraw, T., Mindich, L. and Frangione, B. (1986) Nucleotide sequence of the small double-stranded RNA segment of bacteriophage phi 6:
novel mechanism of natural translational control. J. Virol., 58, 142-151.
21. Mindich, L. and Lehman, J.F. (1979) Cell wall lysin as a component of the bacteriophage ø6 virion. J. Virology, 30, 489-496.
22. Mindich, L. and Lehman, J.F. (1983) Characterization of phi6 mutants that are temperature sensitive in the morphogenetic protein P12.
Virology,127, 438-445.
23. Mindich, L., Nemhauser, I., Gottlieb, P., Romantschuk, M., Carton, J., Frucht, S., Strassman, J., Bamford, D.H. and Kalkkinen, N. (1988)
Nucleotide sequence of the large double-stranded RNA segment of bacteriophage phi 6: genes specifying the viral replicase and transcriptase.
J.Virol., 62, 1180-5.
24. Mindich, L., Qiao, X., Onodera, S., Gottlieb, P. and Strassman, J. (1992) Heterologous recombination in the double-stranded RNA
bacteriophage phi 6. J. Virol., 66, 2605-10.
25. Mindich, L., Sinclair, J.F. and Cohen, J. (1976) The morphogenesis of bacteriophage phi6: particles formed by nonsense mutants. Virology,
75,224-231.
26. Olkkonen, V.M., Ojala, P.M. and Bamford, D.H. (1991) Generation of infectious nucleocapsids by in vitro assembly of the shell protein on to
the polymerase complex of the dsRNA bacteriophage phi 6. J Mol Biol, 218, 569-581.
27. Pirttimaa, M.J., Paatero, A.O., Frilander, M.J. and Bamford, D.H. (2002) Nonspecific nucleoside triphosphatase P4 of double-stranded RNA
bacteriophage phi6 is required for single-stranded RNA packaging and transcription. J Virol, 76.
28. Poranen, M.M., Daugelavicius, R., Ojala, P.M., Hess, M.W. and Bamford, D.H. (1999) A novel virus-host cell membrane interaction.
Membrane voltage-dependent endocytic-like entry of bacteriophage phi6 nucleocapsid. J Cell Biol, 147, 671-82.
29. Poranen, M.M., Paatero, A.O., Tuma, R. and Bamford, D.H. (2001) Self assembly of a viral molecular machine from purified protein and RNA
constituents. Molecular Cell, 7, 845-854.
dsRNA viruses RNA / protein tables: Edited by Peter Mertens and Denis Bamford
30. Roine, E., Raineri, D.M., Romantschuk, M., Wilson, M. and Nunn, D.N. (1998) Characterization of type IV pilus genes in Pseudomonas
syringae pv. tomato DC3000. Mol Plant Microbe Interact, 11, 1048-56.
31. Romantschuk, M. and Bamford, D.H. (1985) Function of pili in bacteriophage phi6 penetration. J. Gen. Virol., 66, 2461-2469.
32. Sinclair, J.F., Tzagoloff, A., Levine, D. and Mindich, L. (1975) Proteins of the bacteriophage phi6. Virology, 75, 685-695.
33. Stitt, B.L. and Mindich, L. (1983) Morphogenesis of bacteriophage phi6: a presumptive viral membrane precursor. Virology, 127, 446-458.
34. Usala, S.J., Brownstein, B.H. and Haselkorn, R. (1980) Displacement of parental RNA strands during in vitro transcription by bacteriophage
phi6 nucleocapsids. Cell, 19, 855-862.
35. Van Etten, J.L., Burbank, D.E., Cuppels, D.A., Lane, L.C. and Vidaver, A.K. (1980) Semiconservative synthesis of single-stranded RNA by
bacteriophage phi6 polymerase. J. Virol., 33, 769-773.