Table 1 Primers and annealing temperature of genes analyzed by
advertisement

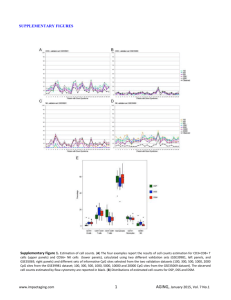
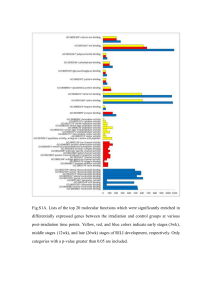
Supplementary Table1: Clinicopathological characteristic of HCC patients included Variable Cohort 1(case numbers) Cohort 2 (case numbers) All cases 18# 31* Age(years), >55:≤55 8:10 13:18 Gender, male/female 16:2 28:3 HBsAg, positive/negative 14:4 25:6 Tumor size(cm), >5:≤5 10:8 14:17 10:8 19:12 Edmondson Grade, I+II: III Patients of group cohort 2 were cohort 1 plus additional 14 cases. # 19 cases were used and 1 of them lost the clinical characteristics. *33 cases were used and 2 of them lost the clinical characteristics. Supplementary Table 2 Primers and annealing temperature of genes analyzed by RT-PCR Primers Temperature (5′to 3′) (℃) Amplification Gene size (bp) F: TGGATGAAAGGGCATTTGAGA PDCD4 55 164 61 324 65 110 64 270 54 201 61 220 64 62 64 90 R: AGCCTTCCCCTCCAATGCTA F: CCGAGTTGGTGATGGTGTGTAC DNMT1 R: AGGTTGATGTCTGCGTGGTAGC F: TATTGATGAGCGCACAAGAGAGC DNMT3A R: GGGTGTTCCAGGGTAACATTGAG F: GACTTGGTGATTGGCGGAA DNMT3B R: GGCCCTGTGAGCAGCAGA F: TTCTTCGTCTGCCGTTCC HBx R: TCGGTCGTTGACATTGCT F: AAAGACCTGTACGCCAACAC ACTB R: GTCATACTCCTGCTTGCTGAT Stem-loop: GTCGTATCCAGTGCAGGGTCC GAGGTATTCGCACTGGATACGACTCAACA miR-21 F:GCCCGCTAGCTTATCAGACTGATG R: GTGCAGGGTCCGAGGT F:CGCTTCGGCAGCACATATACTA U6 R: CGCTTCACGAATTTGCGTGTCA F: TTTAGTTTCGGTTTCGTCGTTAC PDCD4(M) 50 135 55 136 58 194 R: GAAAAATCTCTAACCCTTCTCGC PDCD4(U) F: TTTAGTTTTGGTTTTGTTGTTATGA R: CAAAAAATCTCTAACCCTTCTCACT PDCD4(BGS) F: GGAGTTATTTTTTTATTGAGAGGGG R: CATCTTCAAAAAATTCCCAAAAA Supplementary materials and methods Bisulfite treatment and promoter methylation analysis Genomic DNA was extracted from the cells using the phenol–chloroform method followed by bisulfite modification. Bisulfite genome sequencing was performed as described previously (1). Bisulfite-treated genomic DNA from L02-Vector, L02-HBx #1 and L02-HBx #8 cells were amplified using methylated and unmethylated CDH1 primers as reported previously (2). Primers designed for PDCD4 bisulfite genome sequencing are listed in Supplementary Table 1. Cell lines were treated with 5-Aza (Sigma-Aldrich, St Louis, MO, USA) to determine whether demethylation could restore PDCD4 expression. Briefly, 1×105 cells were treatment with 0, 10, 50 and 100 μM of 5-Aza for 2 days. Wound healing assays Cell migration was assessed by measuring the movement of cells into a scraped area created by a 200-μl pipette tip, and the spread of the wound closure was observed after 24 h. The cells were photographed under a microscope. 1. Tao Q, Huang H, Geiman TM, Lim CY, Fu L, Qiu GH et al. Defective de novo methylation of viral and cellular DNA sequences in ICF syndrome cells. Hum Mol Genet 2002; 11: 2091–2102. 2. Zhang h, xiao w, liang H, Fang d, Yang S, luo Y. Demethylation in the promoter area by the antisense of human dna mtase gene. Zhonghua Zhong liu Za Zhi 2002; 24: 444-447. Supplementary Figure Legends Supplementary Fig. S1. A, HBx expression levels were detected in 10 liver cell lines by RT-PCR. H2O was used as a blank control. A human gastric adenocarcinoma cell line (AGS) was used as negative control. β-actin was used as a loading control. B, The methylation status of the E-cadherin promoter was analyzed using MSP in QSG-7701 cells transfected with HBx and control cells. C, PDCD4 and CDH1 expression was measured by RT-PCR after treatment with 10, 50 and 100 μmol/L 5’-aza for up to 2 days in QSG-7701 cells. β-actin was used as a loading control. D, Mapping of the methylation status of individual CpG site in the PDCD4 promoter by bisulfite genome sequencing in L02-Vector, L02-HBx #1 and L02-HBx #8 cells. The regions spanning the CpG island with 13 CpG sites were analyzed. Methylation status of the PDCD4 promoter was shown. Black color represents the methylated CpG site; achromatic color represents the unmethylated CpG site; every circle represents bisulfite sequencing for 6 random clones. E, In clinical cases with high miR-21 expression (cases in which miR-21 expression was higher than the average expression level in all cases), the rates of PDCD4 down-regulation (T<N) were 63% (5/8) compared with 45% (5/11) of clinical cases with low miR-21 expression (cases which miR-21 expression was lower than average expression level in all cases). Case numbers of every sub-group were represented in the chart. Supplementary Fig. S2. A, Flow cytometry analysis of cell apoptosis. HBx did not significantly influence cell apoptosis when compared to control cells. The values indicate the mean+s.d. for three separate experiments (independent Student’s t-test). B, The effects of PDCD4 on cell migration were detected by wound healing assays and didn’t find significant difference between 7402-PDCD4 and 7402-control cells. Microscopic observation was recorded at 0, 24, and 36 hours after scratching the surface of a confluent layer of cells.