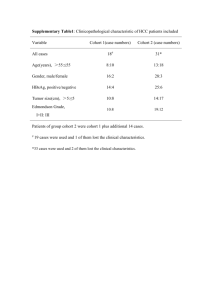
Supplementary Methods (doc 69K)
advertisement

Supplementary Data Normal Colorectal Mucosa Exhibits Sex- and Segment-specific Susceptibility to DNA Methylation at the hMLH1 and MGMT Promoters Menigatti M. et al. 1) Supplementary Information on Experimental Procedures 2) Legends to Supplementary Figures Supplementary Information on Experimental Procedures DNA Extraction, Bisulfite Conversion, Quantitative Methylation Specific PCR and Combined Bisulfite Restriction Analysis Genomic DNA was extracted from the freshly collected samples of normal mucosa and formalin-fixed, paraffin-embedded polyp tissues with the QIAamp DNA Mini Kit (Qiagen) and subjected to sodium bisulfite conversion, as previously described19 with minor modifications. Briefly, DNA (2 g) was diluted in 50 L distilled water and then denatured in 0.3 M NaOH for 20 min at 37°C. After the addition of 30 L of 10 mM hydroquinone (Sigma) and 520 L of 3.46 M sodium bisulfite (Sigma) at pH 5.0, the samples were incubated at 50°C for 16-18 h. The DNA was then purified with the Wizard DNA Clean-Up System (Promega), desulfonated in 0.3 M NaOH for 20 min at 37°C, neutralized with 18 L of 10 M ammonium acetate, ethanol-precipitated, and finally resuspended in 20 L of water. Promoter methylation of hMLH1 and MGMT was analyzed in bisulfite-modified DNA extracted from normal mucosal samples with a quantitative methylation-specific PCR (qMSP) assay. We used allele-specific primers complementary to methylated 1 or unmethylated promoter target sequences. The methyl-specific primers used to assess the MGMT promoter (5’-AGGCGTCGTTCGGTTTGTATCG-3’ [sense] and 5’CGCCGAACCTAAAACAATCTACG-3’ [antisense]) amplified the fragment between positions 703 to 831 (GenBank accession no. X61657). The hMLH1 promoter was analyzed with two sets of allele-specific primers. The first (5’ACGTAGACGTTTTATTAGGGTCGC-3’ [sense] and 5’-TCCGAAAAA CGATAAAACCCTATAC-3’ [antisense]) amplified a segment that was distal to the transcription start site; the second (5’-AACGAATTAATAGGAAGAGCGGATAGCG-3’ [sense] and 5’-CCTCCCTAAAACGACTACTACCCG-3’ [antisense]) amplified a segment proximal to this site. The distal-segment amplicon included nucleotides –716 to –563; that of the proximal segment contained nucleotides –289 to –202 (positions relative to the ATG start site, GenBank accession no. U83845). To quantify input DNA after bisulfite treatment, we amplified a region of the ACTB gene promoter (GenBank accession no. Y00474) free of CpG sites (nucleotides 389 to 521) with the following primers: 5’-GTGGTGATGGAGGAGGTTTAGTA-3’ (sense) and 5’-ACCAATAAAACCTACTCCTCCCTTA-3’ (antisense). Real-time PCR was performed with the Roche LightCycler System and the QuantiTect SYBR Green Kit (Qiagen). The reaction volume (20 L) contained 0.5 M of each primer and 2 L of bisulfite-treated DNA. Amplification conditions were: activation cycle, 95°C for 15min; 55 amplification cycles: 94°C for 15 sec; 58°C (MGMT), 62° (ACTB, hMLH1 distal), or 64° (hMLH1 proximal) for 20 sec; 72°C for 10 sec; followed by a melting-curve step. The high number of amplification cycles was necessary due to the high stringency of the reactions. The specificity of the allele-specific reactions was validated with DNA from the following cancer cell lines, which were kindly provided by the Zurich Cancer Network: SW48 and CO115, 2 colorectal cancer cell lines characterized by full methylation of 2 both gene promoters; A2780/CP70, an ovarian cancer cell line in which the hMLH1 promoter is fully methylated and that of MGMT is unmethylated; and SW620, a colorectal cancer cell line in which the MGMT promoter is fully methylated and that of hMLH1 is unmethylated. Quantitative accuracy was ensured by the inclusion in each run of fully unmethylated control DNA from human peripheral blood leukocytes and GP5D and Caco2 colon-cancer cells (from the Zurich Cancer Network) (Supplementary Figure 1). In a preliminary phase of the study, we also checked the percentages of methylated alleles determined by qMSP against the percentage of unmethylated alleles in the same specimen. The latter was determined in complementary assays with allele-specific primers that annealed to unmethylated sequences in the promoter being assessed (sequences available on request) (Supplementary Figure 2). Standard curves representing serial dilutions of DNA from CO115 cells (described above) were used to define the detection limit of the method and the percentage of methylated alleles in each tissue sample. (Additional details on qMSP assays and their validation are reported in Supplementary Figures 1 and 2.) Promoter methylation in bisulfite-modified DNA from paraffin-embedded polyp tissues was determined by means of combined bisulfite restriction analysis (COBRA). The following primers were used to amplify a 328-bp region of the MGMT promoter: 5’ GAGGATGIGTAGATTGTTTTAG 3’ (sense) and 5’ TCCCAAACACTCACCAAATC 3’ (antisense). For hMLH1, we amplified a 183-bp fragment of the distal region of the promoter (primers: 5’ TTTTTTTAGGAGTGAAGGAGGT 3’ [sense] and 5’ ATAAAACCCTATACCTAATCTATC 3’ [antisense] and a 148-bp fragment of the proximal promoter region (primers: 5’ GAGTTTTTAAAAAIGAATTAATAGGAAGAG 3’ [sense] and 5’ TAAATACCAATCAAATTTCTCAACTCTA 3’ [antisense]). Amplifications were carried out with HotStartTaq DNA polymerase (Qiagen) under 3 the following conditions: denaturing at 95°C for 15 min; amplification in 40 cycles of 94°C for 30 sec, 53°C for 30 sec, and 72°C for 30 sec; extension at 72°C for 10 min. The amplified products were digested with the BstUI restriction enzyme (New England Biolabs) and subjected to electrophoresis in 2.5% agarose gels and ethidium bromide staining. Bisulfite Sequencing of Gene Promoters DNA from several samples of normal mucosa and polyps were subjected to bisulfite sequencing of the hMLH1 and MGMT promoters to determine their methylation status. We used the following primers to amplify a fragment of the MGMT promoter (from positions 666 to 1127, GenBank accession no. X61657) containing 63 CpG sites: 5’-GGGAATTCGTAAATTAAGGTATAGAGTTTTAGG (sense) and 5’-TGTGGATCCCAAACACTCACCAAATC (antisense). A distal fragment of the hMLH1 promoter (-756 to -405 relative to the translation start site, GenBank, U83845) containing 34 internal CpG sites was amplified with the following primers: 5’-TGGGAATTCTTTTTTTAGGAGTGAAGGAGGT (sense) and 5ATAGGATCCTCAAACTCCTCCTCTCC (antisense). Amplifications were carried out with HotStartTaq DNA polymerase (Qiagen) under the following conditions: denaturing at 95°C for 15 min, amplification by 35 cycles of 94°C for 30 sec, 51°C for 30 sec, and 72°C for 1 min, extension at 72°C for 10 min. PCR products were digested with EcoRI and BamHI (New England Biolabs) and subcloned into the pGEM-7Zf(+) Vector (Promega). Individual clones were then sequenced with the PRISM dye terminator cycle sequencing method (Applied Biosystems) (Supplementary Figure 3). Supplementary Figure 4 shows the locations of CpG sites in the hMLH1 and MGMT gene promoters and the regions explored with qMSP, COBRA, and bisulfite sequencing. 4 Legends to Supplementary Figures Supplementary Figure 1: Quantitative methylation-specific PCR (qMSP) assessment of the hMLH1 and MGMT promoters. Reagents were from Qiagen’s QuantiTect SYBR Green kit. (a) Real-time PCR profiles with melting curve (a1) and standard curve (a2) for methylation-specific amplification of the distal region of the hMLH1 promoter. The methylation standard was bisulfite-treated DNA from CO115 colon cancer cells, in which the hMLH1 and MGMT gene promoters are both fully methylated. DNA from the SW48 cell line was used as an additional positive control. The high assay specificity achieved by the use of stringent amplification conditions was verified by the inclusion of fully unmethylated target controls (bisulfite-treated DNA from human peripheral blood leukocytes [PBLs]). (b) Real-time PCR profiles with melting curve (b1) and standard curve (b2) for amplification of a CpG-free region of the ACTB promoter (the internal reference for bisulfite-treated input DNA). Melting curves (a1 and b1) were analyzed to verify the specificity of the PCR reactions (absence of primer dimers and nonspecific products of amplification). Standard curves (a2 and b2) were generated by plotting the threshold cycles (Ct) determined with the second derivative method by the LightCycler software (y-axis) against the log concentrations (x-axis) of the serially diluted standards (i.e., bisulfitetreated DNA from CO115 cells). All reactions were performed in triplicate to provide additional assurance of assay accuracy. (c) Calculation of the percentage of hMLH1 promoter-methylated alleles in a biopsy (example). The numbers of methylated hMLH1 alleles (in the example: 3.278E+03) 5 and bisulfite-treated input alleles (in the example, 2.153E+05) were determined by comparing the Ct of the sample with the standard curves shown in (a2) and (b2), respectively. The percentage of fully methylated hMLH1 alleles in the biopsy was then calculated by dividing the methylated hMLH1 alleles by the bisulfite-treated input alleles and multiplying by 100. Supplementary Figure 2: Validation of the quantitative methylation-specific PCR (qMSP) method. In a preliminary phase of the study, the percentage of alleles presenting promoter methylation in a given DNA sample was verified against the percentage of unmethylated alleles in the same DNA sample determined in complementary assays with allele-specific primers annealing to unmethylated sequences. In the example shown, the level of MGMT promoter methylation in biopsy 30D was determined as described in Supplementary Figure 1. (a) Real-time PCR profiles, melting curves, and standard curves for methylationspecific amplifications of the methylated MGMT promoter sequences (left panel) and the ACTB target sequence (see Supplementary Figure 1) used as an internal reference for bisulfite-treated input DNA (right panel). DNA from CO115 colon cancer cells (in which the MGMT promoter is fully methylated) served as the methylation standard. (b) The DNA sample was then subjected to allele-specific amplifications of the unmethylated MGMT promoter sequences (left panel) and the ACTB sequence used as an internal reference for bisulfite-treated input DNA (right panel). DNA extracted from human peripheral blood leukocytes (PBLs) (in which the MGMT promoter is unmethylated) served as the nonmethylation standard. The percentage of unmethylated MGMT alleles was calculated as the ratio of unmethylated alleles to total bisulfite-treated alleles multiplied by 100. 6 (c) Verification of results: As expected, the sum of the percentages of methylated and unmethylated alleles is close to 100%. Supplementary Figure 3: Quantitative MSP evidence of promoter methylation in DNA from normal-appearing colorectal mucosa was verified by bisulfite DNA sequencing. Upper panel: Sequencing results for a specimen of mucosa displaying methylation of the hMLH1 promoter (distal sequence) involving 10.5% of all alleles. Lower panel: MGMT sequencing in a mucosal sample that exhibited methylation of 7.2% of MGMT alleles. Each row of circles represents the methylation status of the CpG dinucleotides in a single cloned and sequenced allele. Black circles: cytosine methylation; white circles: absence of methylation. Supplementary Figure 4: Schematic depiction of the hMLH1 and the MGMT gene promoters. CpG dinucleotides are represented as short vertical lines. The arrow indicates the putative transcription initiation site. Horizontal bars indicate the regions amplified by qMSP, COBRA, and bisulfite genomic sequencing (dist: distal; prox: proximal). Supplementary Figure 5: Distribution of normal mucosal hMLH1 and MGMT promoter methylation along the colorectum. Histograms show the percentages of subjects with (a) hMLH1 and (b) MGMT promoter methylation involving ≥ 0.1% alleles in each colon segment (C, cecum; TC, transverse colon; SC, sigmoid colon; R, rectum). The frequencies of methylation in the right colon (C and/or TC), left colon (SC and/or R), and in all 4 segments (C & TC & SC & R) were analyzed statistically. Contingency tables were used to calculate expected numbers from the observed numbers, and the resulting distributions were subjected to the Fisher exact test (P 7 values). Detectable methylation in one colon segment significantly increased the likelihood of methylation involving the same promoter in one or two other segments. Supplementary Figure 6. Age- and sex-related trends of promoter methylation in the normal mucosa of polyp-free colons. Scatter charts with data points connected by smoothed lines show percentages of polyp-free women (A, B, n = 35) and men (C, D, n = 34) with detectable methylation (≥ 0.1% alleles) of the hMLH1 (A, C) and MGMT (B, D) promoters across overlapping age-groups. (The statistical power of the analysis shown in this figure was limited by the small number of observations in each age group.) The four colon segments are shown separately, as indicated, with lines connecting the data points to facilitate reading. Figure 3 shows the results of statistical evaluation of the frequency of methylation involving the 2 promoters in the population under 60 years of age and that 60 or over. In the latter group, the prevalence of hMLH1 promoter methylation increased in men and women after the age of 60 (panels A and C). In men, however, there was a decrease in all segments after the age of 70, which was not statistically significant. In women over 60 (panel B), the prevalence of MGMT methylation also increased significantly (see also Figure 3), but it dropped after the age of 70 (significantly in the sigmoid colon [P = 0.022] and rectum [P = 0.028] when age groups 60-70 and 70-80 were compared). In contrast, the prevalence of MGMT methylation did not vary significantly in the colorectum of men (panel D). 8