In Vitro Transcription and Splicing
advertisement
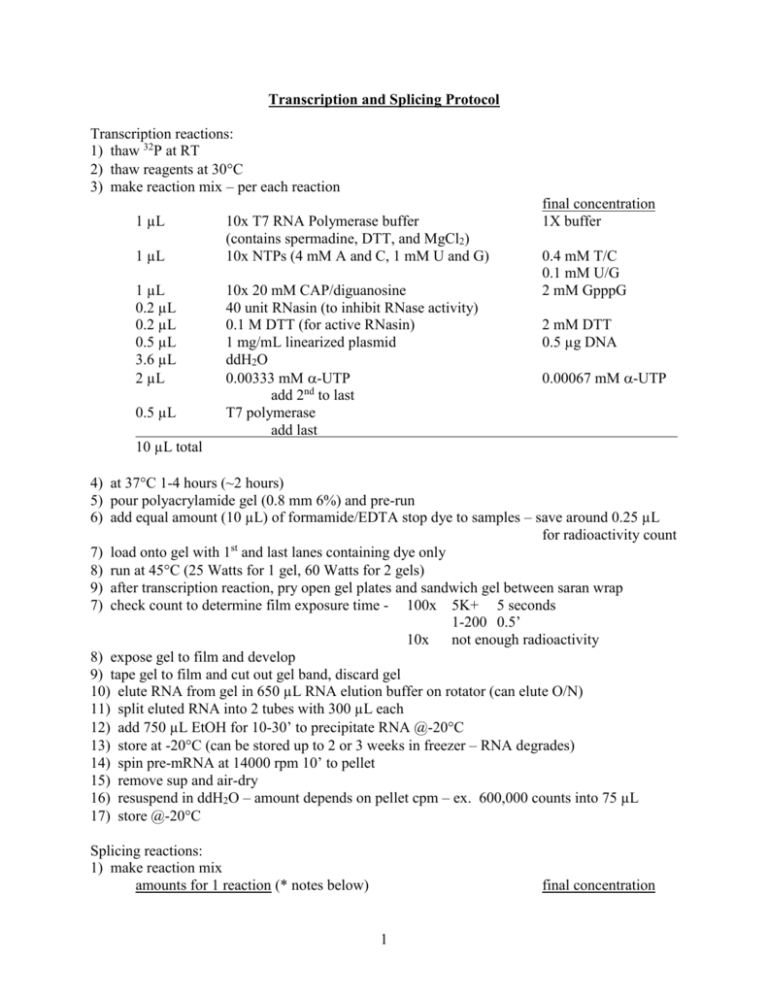
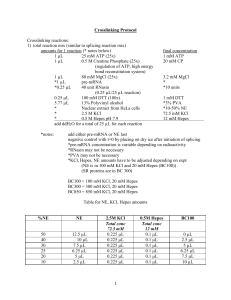
Transcription and Splicing Protocol Transcription reactions: 1) thaw 32P at RT 2) thaw reagents at 30C 3) make reaction mix – per each reaction 1 µL 1 µL 1 µL 0.2 µL 0.2 µL 0.5 µL 3.6 µL 2 µL 0.5 µL 10x T7 RNA Polymerase buffer (contains spermadine, DTT, and MgCl2) 10x NTPs (4 mM A and C, 1 mM U and G) 10x 20 mM CAP/diguanosine 40 unit RNasin (to inhibit RNase activity) 0.1 M DTT (for active RNasin) 1 mg/mL linearized plasmid ddH2O 0.00333 mM -UTP add 2nd to last T7 polymerase add last final concentration 1X buffer 0.4 mM T/C 0.1 mM U/G 2 mM GpppG 2 mM DTT 0.5 µg DNA 0.00067 mM -UTP 10 µL total 4) at 37C 1-4 hours (~2 hours) 5) pour polyacrylamide gel (0.8 mm 6%) and pre-run 6) add equal amount (10 µL) of formamide/EDTA stop dye to samples – save around 0.25 µL for radioactivity count st 7) load onto gel with 1 and last lanes containing dye only 8) run at 45C (25 Watts for 1 gel, 60 Watts for 2 gels) 9) after transcription reaction, pry open gel plates and sandwich gel between saran wrap 7) check count to determine film exposure time - 100x 5K+ 5 seconds 1-200 0.5’ 10x not enough radioactivity 8) expose gel to film and develop 9) tape gel to film and cut out gel band, discard gel 10) elute RNA from gel in 650 µL RNA elution buffer on rotator (can elute O/N) 11) split eluted RNA into 2 tubes with 300 µL each 12) add 750 µL EtOH for 10-30’ to precipitate RNA @-20C 13) store at -20C (can be stored up to 2 or 3 weeks in freezer – RNA degrades) 14) spin pre-mRNA at 14000 rpm 10’ to pellet 15) remove sup and air-dry 16) resuspend in ddH2O – amount depends on pellet cpm – ex. 600,000 counts into 75 µL 17) store @-20C Splicing reactions: 1) make reaction mix amounts for 1 reaction (* notes below) final concentration 1 1 µL 1 µL 25 mM ATP (25x) 0.5 M Creatine Phosphate (25x) (regulation of ATP, high energy bond reconstitution system) 1 µL 80 mM MgCl2 (25x) *1 µL pre-mRNA *0.25 µL 40 unit RNasin (0.25 µL/25 µL reaction) 0.25 µL 100 mM DTT (100x) 5.77 µL 13% Polyvinyl alcohol * Nuclear extract from HeLa cells * 2.5 M KCl * 0.5 M Hepes pH 7.9 add ddH2O for a total of 25 µL for each reaction *notes: 1 mM ATP 20 mM CP 3.2 mM MgCl * *10 units 1 mM DTT *3% PVA *10-50% NE 72.5 mM KCl 12 mM Hepes add either pre-mRNA or NE last negative control with t=0 by placing on dry ice after initiation of splicing *pre-mRNA concentration is variable depending on radioactivity *RNasin may not be necessary *PVA may not be necessary *KCl, Hepes, NE amounts have to be adjusted depending on expt (NE is in 100 mM KCl and 20 mM Hepes (BC100)) (SR proteins are in BC 300) BC100 = 100 mM KCl, 20 mM Hepes BC300 = 300 mM KCl, 20 mM Hepes BC850 = 850 mM KCl, 20 mM Hepes Table for NE, KCl, Hepes amounts %NE NE 50 40 30 25 20 10 12.5 µL 10 µL 7.5 µL 6.25 µL 5 µL 2.5 µL 2.5M KCl Total conc 72.5 mM 0.225 µL 0.225 µL 0.225 µL 0.225 µL 0.225 µL 0.225 µL To determine SR protein stock molarity: use concentration (mg/mL = g/L) convert g to Da (1 Da = 1 g) (g/L) / (g/mol) = Molar 0.5M Hepes Total conc 12 mM 0.1 µL 0.1 µL 0.1 µL 0.1 µL 0.1 µL 0.1 µL BC100 0 µL 2.5 µL 5 µL 6.25 µL 7.5 µL 10 µL ex. 0.4 mg/mL = 0.4 g/L 45 kDa prot = 45000 g (g/mol) (0.4 g/L) / (4500 g/mol) 2 = 9 x 10-6 M = 9 µM NE Table for BC100, BC300, NE, and SR protein amounts BC100 SR protein BC300 0.5M Hepes 10 µL 1.5 µL 0.14 µL 2.5M KCl 0.15 µL 2) at 30C for 90’ 3) make proteinase K digestion mix – for each 25 µL reaction 100 µL 2x pK buffer 75 µL ddH2O 2.5 µL 10 mg/mL proteinase K 1.5 µL glycogen (ppt carrier) 4) add 175 µL of mix to each reaction 5) at 37C 12-15’ 6) add 200 µL phenol:chloroform 1:1 to extract RNA 7) vortex 5-10 seconds 8) spin at 14000 rpm 2-5’ 9) remove and save ~200 µL sup 10) add ~500 µL ethanol to ppt RNA 11) place at -20C for at least 10’ 12) spin at 14000 rpm 10’ to pellet 13) remove sup 14) resuspend in 7 µL formamide/EDTA stop buffer 15) run on polyacrylamide gel (0.7 mm 6% and/or 10% 16) after gel run, can vacuum dry onto paper or sandwich gel between saran wrap 17) expose to film with intensifying screen at -80C O/N or expose to phosphor screen 3 For polyacrylamide gels: 1) clean plates with ethanol 2) set-up plates with clamps and spacers 3) make gel mix: 4) pour gel at angle 5) allow to polymerize 6) can pre-run to get rid of free radicals in Temed For 5% polyacrylamide gels: 1x TBE 7 M Urea 20% polyacrylamide/bis 19:1 7 M Urea 1x TBE 10% APS Temed 20 mL 15 mL 5 mL 30 mL 22.5 mL 7.5 mL 40 mL 30 mL 10 mL 200 L 40 L 300 µL 60 µL 400 µL 80 µL 20 mL 14 mL 6 mL 30 mL 21 mL 9 mL 40 mL 28 mL 12 mL 200 L 40 L 300 µL 60 µL 400 µL 80 µL 20 mL 12 mL 8 mL 30 mL 18 mL 12 mL 40 mL 24 mL 16 mL 200 L 40 L 300 µL 60 µL 400 µL 80 µL 20 mL 10 mL 10 mL 30 mL 15 mL 15 mL 40 mL 20 mL 20 mL 200 L 40 L 300 µL 60 µL 400 µL 80 µL For 6% polyacrylamide gels: 1x TBE 7 M Urea 20% polyacrylamide/bis 19:1 7 M Urea 1x TBE 10% APS Temed For 8% polyacrylamide gels: 1x TBE 7 M Urea 20% polyacrylamide/bis 19:1 7 M Urea 1x TBE 10% APS Temed For 10% polyacrylamide gels: 1x TBE 7 M Urea 20% polyacrylamide/bis 19:1 7 M Urea 1x TBE 10% APS Temed 4