Crosslinking Protocol

Crosslinking Protocol
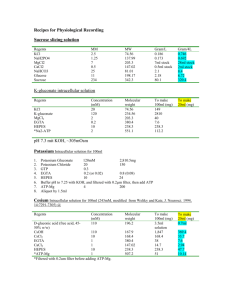
Crosslinking reactions:
1) total reaction mix (similar to splicing reaction mix) amounts for 1 reaction (* notes below)
1 µL
1 µL
25 mM ATP (25x)
0.5 M Creatine Phosphate (25x)
(regulation of ATP, high energy
1 µL
*1 µL bond reconstitution system)
80 mM MgCl (25x) pre-mRNA
*0.25 µL
0.25 µL
5.77 µL
40 unit RNasin
(0.25 µL/25 µL reaction)
100 mM DTT (100x)
13% Polyvinyl alcohol
*
*
Nuclear extract from HeLa cells
2.5 M KCl
* 0.5 M Hepes pH 7.9 add ddH
2
O for a total of 25 µL for each reaction
*notes: final concentration
1 mM ATP
20 mM CP
3.2 mM MgCl
*
*10 units
1 mM DTT
*3% PVA
*10-50% NE
72.5 mM KCl
12 mM Hepes add either pre-mRNA or NE last negative control with t=0 by placing on dry ice after initiation of splicing
*pre-mRNA concentration is variable depending on radioactivity
*RNasin may not be necessary
*PVA may not be necessary
*KCl, Hepes, NE amounts have to be adjusted depending on expt
(NE is in 100 mM KCl and 20 mM Hepes (BC100))
(SR proteins are in BC 300)
BC100 = 100 mM KCl, 20 mM Hepes
BC300 = 300 mM KCl, 20 mM Hepes
BC850 = 850 mM KCl, 20 mM Hepes
Table for NE, KCl, Hepes amounts
%NE
50
40
30
25
20
10
NE
12.5 µL
10 µL
7.5 µL
6.25 µL
5 µL
2.5 µL
2.5M KCl
Total conc
72.5 mM
0.225 µL
0.225 µL
0.225 µL
0.225 µL
0.225 µL
0.225 µL
0.5M Hepes
Total conc
12 mM
0.1 µL
0.1 µL
0.1 µL
0.1 µL
0.1 µL
0.1 µL
BC100
0 µL
2.5 µL
5 µL
6.25 µL
7.5 µL
10 µL
1
To determine SR protein stock molarity: use concentration (mg/mL = g/L) convert g to Da (1 Da = 1 g)
(g/L) / (g/mol) = Molar ex. 0.4 mg/mL = 0.4 g/L
45 kDa prot = 45000 g (g/mol)
(0.4 g/L) / (4500 g/mol)
= 9 x 10
= 9 µM
Table for BC100, BC300, NE, and SR protein amounts
-6 M
0.5M Hepes 2.5M KCl
0.14 µL 0.15 µL
NE
10 µL
BC100 SR protein
1.5 µL
BC300
2) make mix and preincubate @30 C for approximately 25-30’
3) Xlink for 10’ in 0.6 mL tube against UV bulb (on ice)
(Xlink conditions are variable)
4) add 1 L (10 mg/mL) RNase
5) @30 C for 10’
6) add 10 L 2x SDS-PAGE buffer
7) boil samples 5’
8) run on SDS-PAGE 100 V 60’
9) after gel run, vacuum dry onto blot paper
10) expose to phosphorimager
2
For 10% SDS-PAGE:
1) clean plates with ethanol
2) set-up apparatus
3) make resolving gel mix: water
30% polyacrylamide (29:1)
1.5 M Tris pH 8.8
10% SDS
10% APS temed
4) pour gel at angle
5) use 0.01% SDS or butanol to keep top of gel even
6) allow to polymerize
7) get rid of 0.01% SDS or butanol with blot paper
8) make stacking gel mix: water
30% polyacrylamide (29:1)
1 M Tris pH 6.8
10% SDS
10% APS temed
9) pour stacking gel, add gel comb
10) allow to polymerize
~5 mL
1.9 mL
1.7 mL
1.3 mL
50 L
50 L
8 L
~1 mL
680 L
170 L
130 L
10 L
10 L
10 L
3