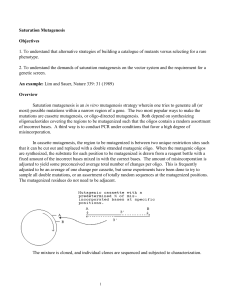
09..Directed mutagenesis
advertisement

p.1
09. Directed mutagenesis
- 'reverse genetics' :
classical genetics : introduce mutations “randomly” to create a phenotype
=> locate the mutated site
'reverse' genetics : create a mutation at a well-determined position
=> analyse for an eventual phenotypic effect
- three basic approaches: (1) cassette mutagenesis (2) primer extension (3) PCR
Cassette mutagenesis
- excise a fragment (restrictions sites!) + replace by a synthetic fragment
- FOR
- high efficiency
- multiple exchanges are possible, including degenerations
- AGAINST
- flanking "unique" cleavage sites necessary
- synthetic capacity should be high enough
- use of overlapping sets of oligonucleotides is possible (see chemical synthesis)
- when complex degenerations: make 2nd strand by a fill-in reaction
(hybridisation kinetics !!!)
Mismatch primer extension : oligonucleotide-directed, mismatch-dependent
- requires single stranded template (may be partially ss)
- M13- or phasmid clone
- preparation of gapped-duplex
- requires mismatch oligonucleotide (chemical synthesis)
- position of mismatch in oligonucleotide onto template is critical
G. Volckaert
- 3'-effect (repair)
=> choice of polymerase !
- 5'-effect (displacement)
=> choice of polymerase !
Directed mutagenesis
12/02/2016
p.2
- possibilities: point mutations, multiple point mutations, insertion, deletion
('sticky feet'-mutagenesis)
- efficiency is low unless special measures are taken
- several factors involved
- transformation by heteroduplex + original template
- repair mechanisms in E.coli (GATC !)
=> use of mutL, mutS, mutH strains
- selection of mutant strand : some examples
- method of Eckstein :
- in vitro fixation of the mutation
- use of phosphorothioates (S-dCTP)
- restriction enzyme versus S-modification
- method of Kunkel :
- in vivo selection of mutant
- "doped" template strand with uridylate
- double mutant
dut (dUTPase)
ung (uracil-N-glycosidase)
- gapped duplex method, with mismatched complementary strand
- transformation to a non-suppressor strain (Su-)
- parental DNA requires amber suppressor
- gapped duplex method, with alternating amber mutants :
- Apam => ApR and CmR => Cmam, and vice versa
- transformation to a Su- strain
- in vivo selection with changing selection marker :
- inclusion of a selection primer to change the bla gene
- mutant bla gene confers resistance to ceftazidime + ampicillin
- non-mutant transformants do not survive with ceftazidime
- T4 polymerase provides efficient 'linkage' between the primers
(no 5'-3' exo, no strand displacement)
In the cassette- and mismatch-dependent mutagenesis approaches, modified
oligonucleotides or modifications in the mismatch primer can be used, e.g. using
inosine.
G. Volckaert
Directed mutagenesis
12/02/2016
p.3
PCR-based site-specific mutagenesis
- elimination of the wild-type template by amplification
- mismatch primer on target position
=> "back-priming” (in 2nd cycle) fixes the change
- cfr. cassette mutagenesis, but the regions (fragments) can be much larger.
- restriction site "in the neighborhood" is required, but may be nevertheless
at a reasonbly larger distance (e.g. 25 nt)
- the need for a restriction site can be circumvented in an approach reminiscent
of SOE.
- 'megaprimer' mutagenesis : two-step procedure, in which the early amplicon
product becomes the primer in subsequent amplification
- by inverse PCR : if the vector is sufficiently small
- linear product
- circularisation required
- cleavage position again useful
- 'tailed' primers can also lead to insertional mutagenesis
- other alternatives :
- use of EarI to efficiently recircularize (inverse PCR)
- example of counterselection with DpnI :
- DpnI cleaves only Dam-methylated DNA (at least in one strand)
- template is Dam-methylated
- mutant strand and PCR product are not methylated
- DpnI digestion before transformation eliminates original DNA
(including the molecules with one strand methylated)
PCR methods have become the major approach for directed mutagenesis
- FOR :
- efficiency nearly 100% (under optimal conditions)
- simplicity
G. Volckaert
Directed mutagenesis
12/02/2016
p.4
- AGAINST :
- product is linear: insertion in a vector or circularisation
before cloning is required
- risk of extra mutations (in the remainder of the amplicon :
=> requires sequencing!)
=> choice of polymerase is important
Detection (confirmation)
not at exam
- physically: creation or removal of cleavage sites (restriction analysis)
(reverse-translation analysis to find potential manipulations)
- sequencing: extra primer required at short distance from the mismatch primer
- hybridisation: +/- analysis with mismatch primer
G. Volckaert
Directed mutagenesis
12/02/2016