Saturation Mutagenesis

Saturation Mutagenesis
Objectives
1. To understand that alternative strategies of building a catalogue of mutants versus selecting for a rare phenotype.
2. To understand the demands of saturation mutagenesis on the vector system and the requirement for a genetic screen.
An example: Lim and Sauer, Nature 339: 31 (1989)
Overview
Saturation mutagenesis is an in vitro mutagenesis strategy wherein one tries to generate all (or most) possible mutations within a narrow region of a gene. The two most popular ways to make the mutations are cassette mutagenesis, or oligo-directed mutagenesis. Both depend on synthesizing oligonucleotides covering the regions to be mutagenized such that the oligos contain a random assortment of incorrect bases. A third way is to conduct PCR under conditions that favor a high degree of misincorporation.
In cassette mutagenesis, the region to be mutagenized is between two unique restriction sites such that it can be cut out and replaced with a double stranded mutagenic oligo. When the mutagenic oligos are synthesized, the substrate for each position to be mutagenized is drawn from a reagent bottle with a fixed amount of the incorrect bases mixed in with the correct bases. The amount of misincorporation is adjusted to yield some preconceived average total number of changes per oligo. This is frequently adjusted to be an average of one change per cassette, but some experiments have been done to try to sample all double mutations, or an assortment of totally random sequences at the mutagenized positions.
The mutagenized residues do not need to be adjacent.
The mixture is cloned, and individual clones are sequenced and subjected to characterization.
1
Non PCR mutagenesis methods.
Site Specific Mutagenesis methods
M13
This example from Winter et al., Nature 299: 756-758 (1982), yielded a surprisingly successful result from a very simple strategy. The gene to be mutagenized was isolated from a thermophilic bacterium, Bacillus stearothermophilus . Simply placing it on an M13 vector was sufficient to express the transgene at 50% of soluble protein from its own expression signals. The E. coli enzyme (along with most other E. coli proteins) denatures and precipitates at high temperature, whereas the transgenic protein does not. Therefore, a heat precipitation step sufficed to greatly purify the protein and eliminate the endogenous activity.
Single stranded M13 + strand carrying the target gene was primed with a mutagenic oligonucleotide, extended, ligated, and used to transfect the E. coli host. The duplex will have a base mismatch, and the E. coli repair systems will fix the resulting heteroduplex by repairing some clones to the wild type sequence, and some to the directed mutation.
Figure modified from Winter et al., Nature 299: 756-758 (1982).
In this instance, they screened to distinguish mutant from wild type plaques by hybridization to the mutagenic oligonucleotide under stringent conditions where the one base mismatch would preclude hybridization. One might expect 50% of clones to contain the mutant (as was the case in this experiment), but more typically, one only gets about 1% mutants. There are two general causes of this problem. 1)
Molecules that are not completely extended and ligated can be repaired after entry into E. coli . The gap will be filled by pol I, which will use its 5' exonuclease to remove the mutagenic oligonucleotide. This problem can be addressed by destroying the gapped molecules with S1 nuclease, or by physically
2
separating the closed circular molecules, or by improving the polymerization to eliminate gapped molecules. 2) The repair systems can read the methylation patterns on the heteroduplex, and distinguish the original template (methylated) from the in vitro synthesized strand. They then preferentially fix the heteroduplex back to the wild type. The repair systems can be redirected to fix the heteroduplex towards the mutant strand by a genetic trick referred to as the "Kunkel method" (Kunkel, T.A., Proc. Natl. Acad.
Sci. USA 82:488 (1982)).
Kunkel method: When E. coli carries a mutation named dut there is a buildup of dUTP in the cells and some uracil is incorporated into newly synthesized DNA. There is a repair system that detects the uracil and removes that section of DNA leading to resynthesis of that region. If one arranges to incorporate uracil into the wild type strand, then this system will cause the wild type strand to be recopied from the mutant strand after entry into the host, leading to repair of the heteroduplex towards the mutant sequence. Uracil is incorporated in the wild type strand by growing the template in a dut ung host. The ung mutations knocks out the repair system in that host, so that the incorporated uracil is never removed.
This trick also provides some protection against transformation by gapped molecules, because cleavage of the uracil containing strand in the gapped region cannot be repaired, so those molecules are destroyed.
Alternative methods to prefer the mutated strand over the parental template include using a MutS background (blocks mismatch repair) and cleaving with the methylation-dependent restriction enzyme
Dpn I.
Phagmids
The Kunkel method is usually carried out on phagmids. These are plasmids that additionally carry the M13 + strand origin of replication. The plasmid could be an expression vector with all the features required for production/characterization of the target molecule. In addition, when grown in the dut - ung host in the presence of an M13 helper phage, a single strand, uracil-substituted circle is produced and packaged in the M13 capsid from which it can be purified ready to use for mutagenesis.
Co-mutation .
If two different mutagenic primers are used on the same single-stranded template, then both mutations are incorporated at once. The initial use of co-mutation was to create a selection for prodicts containing the mutations. Using comutation, one can incorporate one of the mutations in an antibiotic resistance gene, to allow direct selection for mutagenized clones. This will generally require a specialized vector with two antibiotic resistance genes. Promega has a system where mutagenic primers alter the amp gene to confer resistance to additional beta-lactam antibiotics. This system can be used on any vector that has the amp resistance marker. Along the same lines, one can add or remove a unique restriction site with the second mutation, and use it to discriminate between mutated and unmutated clones. One can also make the second mutation either introduce or remove an amber (nonsense) mutation.
For example, one could make the amp resistance gene become conditional on a suppressor.
The reason to revert to this older technology for saturation mutagenesis, is that the comutation strategy can allow simultaneous saturation at codons that are far removed in the sequence.
3
Cataloging versus Screening
If the sought-after phenotype is expected to be found in a major proportion of the mutants, then one often sequences the candidates at random, and then tests them for the phenotype one at a time. The negatives are compiled as well as the positives, thus one can potentially tell that all of the possible mutations have actually been examined.
If the sought-after phenotype is expected to be rare among a very large number of possible mutations, then one would conduct the phenotypic examination first as a genetic screen, and then just sequence the clones that pass the screen.
Demands on the vector host system
One wants to do the mutagenesis, screening, and sequencing in the same vector. Therefore, the vector has to be well designed from the beginning of the experiment. If the target sequence is a cis control sequence, then it should be fused to an easily screened reporter gene. If the target is an element of a protein sequence, then the activity of the protein should be monitored as directly as possible in a genetic screen.
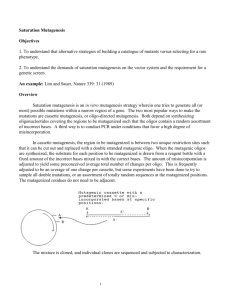
Hydrophobic Core of the Lambda Repressor
The following is an example of a superbly designed experimental system. The lambda repressor
(C
I
) was subjected to saturation mutagenesis of the set of amino acids composing the hydrophobic core.
These were 7 noncontiguous residues that were mutated within the context of a 102 residue cassette as in the figure above. The degree of misincorporation was such that variants at between 1 and 4 of the 7 amino acids were recovered.
Selection for function of the repressor was done by infecting the cells with a strain of lambdaphage carrying a mutant repressor gene. Consequently, clones expressing nonfunctional repressor were killed by the lambda phage, whereas those expressing functional repressor were able to repress the lytic functions of the phage and survive.
The mutagenized repressors were driven by an inducible tac promoter. Some were able to repress the incoming lambda phage when expressed at the basal level, and others could only successfully repress if the tac promoter was induced. Therefore, the mutants were divided into three classes: totally functional
(similar to the wild type); partially functionally (requires over expression); and nonfunctional. 30-100 mutants of each class were sequenced.
The scientific result of this experiment was that you can make a lot of changes in the hydrophobic core as long as the residues are still hydrophobic and the total volume of the core is not much changed.
The technical aspect of the experiment that I would like to emphasize is the placement of the selection before the sequencing. Only about 1% of the mutants survived the screen. If they had sequenced at random, and then tested the phenotype later, they would have obtained most of the data from
4
the nonfunctional set. The application of the selection first allowed them to pick similar numbers of mutants to sequence from each set.
One demand imposed on the vector in this experiment is that you make the mutations directly in the expression vector. That means that they had to do some engineering in advance to create the cassette in an appropriate expression vector. Another demand is that expression from the vector is at a suitable level to conduct a genetic test. In this experiment, they used the fact the over expression can alter the phenotype to their advantage. Ignoring this fact could lead to some seriously irreproducible results (such as reporting mutants as wild type and then finding out that they fail to function in other assays).
Now imagine doing such an experiment only conducting the screen in a eucaryotic cell. The demands on the vector will define what I mean by a true shuttle vector. If you tried to do this with an acute transfection in COS cells, you could never separate the positives from the negatives, because you never get clones in that assay, just a mixed population of independently transfected cells. So you would be constrained to sequence each candidate first while it was still in E. coli , and then test them one at a time in independent transfection experiments.
A true shuttle vector would maintain the mutants in the eucaryotic cell long enough to be separated into individual clones and then subjected to a genetic test or screen or selection. The ones that pass should then be able to be shuttled back into E. coli for sequencing. True shuttle vectors exist for yeast. Some vectors exist for mammalian cells that approach these capabilities, but all of them have problems. One common problem is that reliable tightly controlled promoters are not available, such that you can keep the transgene turned off until you are ready to do the screen. This leads to the risk of inadvertently conducting a selection during clonal expansion based on toxicity of the transgene. A second problem is that many of the vectors integrate into the genome. This allows for multiple integrations that will not segregate apart the way that plasmids do in bacteria.
Two technical advances exist that may eventually substitute for the shuttle vector in a eucaryotic saturation mutagenesis experiment. One is the Fluorescence Activated Cell Sorter (FACS) that could conduct a screen one cell at a time, removing the requirement to grow the transfected population up as colonies. The second is PCR which may allow recovery of the molecules from individual sorted cells.
Study problem
1. How could you carry out saturation mutagenesis of the SV-40 enhancer? (SV-40 is a virus that infects mammalian cells).
2. How could you use the fact that human ras will complement yeast ras to carry out saturation mutagenesis looking for other mutations (besides val 119) that produce an activated ras? (Yeast ras functions in a pathway that signals vegetative growth under conditions of good nutrition and sporulation under conditions of poor nutrition).
3. What would you need to recover a collection of stability mutants in rhodanese by saturation mutagenesis? (Rhodanese transfers a sulfur to cyanide).
5