GRININGER_SUPPORT_REV
advertisement

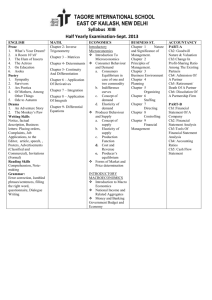
Supporting information for Electrochemical Switching of the Flavoprotein Dodecin at Gold Surfaces Modified by Flavin-DNA Hybrid Linkers by Martin Grininger, Gilbert Nöll, Sibylle Trawöger, Eva-Kathrin Sinner, Dieter Oesterhelt S1 Experimental Part Synthesis of the flavins Scheme S1. Reaction scheme leading to flavins with alkyl amino chains. 1) H pyridine, Na2CO3 Cl + H2N n n NHBoc n = 4: yield: 47% NO2 NO2 H 2) NHBoc N H NHBoc N Pd/C, acetic acid NHBoc N n n H2, MeOH NO2 NH2 NHBoc H 3) H N O NHBoc N N NH O n n O N O NH2 NH N H3BO3 O O n = 4: yield over two steps: 48% O 4) NHBoc F3C + NH3 O n n N N O NH N N TFA n = 4: yield: 85% N O NH N O O The reaction scheme (Scheme S1) leads to flavin derivatives bearing an alkyl amino group at N(10) and was carried out analogously to the synthesis of the homologue hexyl amino derivative.1 The preparation required prior protection of the amino group finally used for further linkage. The yields are not optimized. Highest yields were obtained at medium chain length (n = 4 and 5). Shorter chain lengths (n = 2 and 3) resulted in lower solubility of the boc-protected flavins whereas in the case of longer chain lengths (n = 6 and 8) the nucleophilic aromatic substitution at 1-Chloro-2-nitro-4,5-dimethylbenzene was less effective. Representatively, the synthesis of N(10)-[4-(aminobutyl] flavin • TFA (CofC4 • TFA) is described below. S2 1-[(tButyl)oxycarbonylaminobutyl]-2-nitro-4,5-dimethylbenzene The mono-Boc protected butylene diamine (4.86 g; 25.8 mmol) and 1-Chloro-2-nitro-4,5dimethylbenzene (5.6 g; 30.1 mmol) were dissolved in pyridine (30 ml). After addition of 3 g Na2CO3 the reaction mixture was stirred for 4-5 d at 90-100 °C and subsequently concentrated in vacuo. The residual material was dissolved in CHCl3 and washed with H2O. After evaporation of the solvent the product was purified by flash-chromatography (silica gel, solvent: CH2Cl2).Yield: 4.1 g; 47 %; 12.2 mmol, m.p.: 108-110 °C, C17H27N3O4; 337.42 g/mol, EA: calc.: C: 60.51, H: 8.07, N: 12.45; found: C: 60.49, H: 7.68, N: 12.15; CI-MS (NH3): MH+: 340(2%), 339(17%), 338(100%), additional peaks at: 282(28%), 308(26%), 282(28%), 281(12%), 264(39%), 238(25%), 234(43%). H N 6 5 4 1 3 a O d b c N H O 2 NO2 1H-NMR (400 MHz, CDCl3): δ = 1.45(9H, s, tBu), 1.55-1.68(2H, m, Hc), 1.70-1.85(2H, m, Hb), 2.18(3H, s, CH3 at C4), 2.27(3H, s, CH3 at C5), 3.17-3.25(2H, m, Hd), 3.31(2H, t, J = 7.0 Hz, Ha), 4.55(1H, s, br, NH), 6.62(1H, s, H6), 7.92(1H, s, H3) determination was done by HMBC, HSQC and COSY. 13C-NMR (100,6 MHz, CDCl3): = 18.6(CH3 at C4), 20.7(CH3 at C5), 26.4(Cb), 27.8(Cc), 28.4((CH3)3), 40.2(Cd), 42.6(Ca), 79.3(C(CH3)3), 114.1(C6), 124.4(C4), 126.5(C3), 129.8(C2), 144.1(C1), 147.3(C5), 156.0(N-C=O); determination was done by HMBC, HSQC and COSY. 1-[4-(tButyl)oxycarbonylamino-butyl]-2-amino-4,5-dimethylbenzene A suspension of 10% Pd/C catalyst (50 mg) in acetic acid (3 ml) was added to a orange-red solution of 1-[(tButyl)oxycarbonylaminobutyl]-2-nitro-4,5-dimethylbenzene (4.1 g, 12.2 mmol) in methanol (50 ml). The suspension was stirred at 60 °C in a H2 atmosphere until it became colorless (about 24 h). The reaction mixture was filtered through celite under inertgas atmosphere. The product is very sensitive against oxidation. Therefore the next reaction step was done immediately without further purification. C17H29N3O2; 307.44 g/mol N(10)-[4-(tButyl)oxycarbonylamino-butyl] flavin Alloxan monohydrate (6.0 g, 37.4 mmol) and boric acid (9.0 g, 148 mmol) were added to the filtrate containing 1-[4-(tButyl)oxycarbonylamino-butyl]-2-amino-4,5-dimethylbenzene under inertgas atmosphere and stirred for 24 h at room temperature. The reaction mixture was diluted with CHCl3 (300 ml) and extracted with water (3 times 100 ml). The organic phase was separated and dried with magnesium sulphate. After removing the solvent the yellow-orange product was purified by flashchromatography on silica gel (CH2Cl2/MeOH 100:2). Yield (over two steps): 2.40 g; 48 %; 5.80 mmol; S3 m.p.: pyrolysis at 236 °C; C21H27O4N5; 413.48 g/mol; EA: calc.: C: 61.00, H: 6.58, N: 16.94; found: C: 61.24, H: 6.63, N: 15.59, ESI-MS : MH+: 414(100%); 415(25%); MNa+: 436(15%) O N O d c b a 9 10 N N 1 8 7 N 5 6 4 O 2 NH 3 O 1H-NMR (600 MHz, DMSO-d6): = 1.38(9H, s, tBu), 1.54(2H, m, Hc), 1.71(2H, m, Hb), 2.40(3H, s, CH3 at C7), 2.52(3H, s, CH3 at C8), 2.98(2H, m, Hd), 4.57(2H, m, Ha), 6.83(1H, m, NH), 7.80(1H, s, H9), 7.89(1H, s, H6), 11.29(1H, s, NH-Ar). determination was done by HMBC, HSQC and COSY. 13C-NMR (150,9 MHz, DMSO-d6): = 18.7(CH3 at C7), 20.6(CH3 at C8), 24.0(Cb), 26.7(Cc), 28.2((CH3)3), 39.3(Cd), 43.9(Ca), 77.4(C(CH3)3), 116.1(C9), 130.7(C9a), 131.0(C6), 133.7(C5a), 135.7(C7), 137.1(C4a), 146.5(C8), 150.0(C10a), 155.60(N-C=O), 155.63(C2), 159.9(C4). N(10)-[4-(aminobutyl] flavin • TFA, CofC4 • TFA A solution of the Boc-protected flavin derivative (2.4 g; 5.80 mmol) was stirred in TFA/water (30 ml, 95:5) for 3 h at room temperature. The reaction solution was evaporated in vacuo and the product was precipitated through addition of diethyl ether. CofC4 was obtained as the TFA salt in form of a yellow powder. Further purification was done by precipitation (the product was dissolved in a small amount of MeOH/CH2Cl2 and poured into a stirred solution of Et2O). Yield: 2.10 g; 4.91 mmol; 85 %; m.p.: pyrolysis at 245 °C; C18H20F3N5O4; 427.38 g/mol; EA: calc.: C:50.59, H:4.72, N: 16.39; found: C:50.10, H:4.89, N: 16.26; EI-MS(PI) : M+(amine): 313(17%), 314(2%), 242(38%), 171(18%), 279(38%), 293(60%); LSI-MS(PI): MH+(amine): 314(55%), 315(100%), 316(40%). + NH3 d c O b a 9 10 N 6 N 5 N 8 7 F -O 4 1 F F O 2 NH 3 O S4 1H-NMR (400 MHz, DMSO-d6): = 1.68(2H, m, Hc), 1.83(2H, m, Hb), 2.41(3H, s, CH3 at C7), 2.52(3H, s, CH3 at C8), 2.90(2H, m, Hd), 4.65(2H, m, Ha), 7.76(3H, s, br, NH3+), 7.83(1H, s, H9), 7.92(1H, s, H6), 11.35(1H, s, NH). determination was done by HMBC, HSQC and COSY. 13C-NMR (100,6 MHz, DMSO-d6): = 19.2(CH3 at C7), 21.1(CH3 at C8), 24.0(Cb), 24.5(Cc), 38.9(Cd), 43.7(Ca), 116.6(C9), 117.8(q, JCF3-19F = 299.63 Hz, CF3), 131.1(C9a), 131.6(C6), 134.3(C5a), 136.4(C7), 137.6(C4a), 147.1(C8), 150.6(C10a), 156.1(C2), 158.3(q, JCOO(CF3)-19F = 30.44 Hz, COO), 160.4(C4). determination was done by HMBC, HSQC and COSY. Analytical data of other artificial flavins: N(10)-[4-(aminoethyl] flavin • TFA, CofC2 • TFA See also refs. 2,3 C16H16F3N5O4; 399.33 g/mol; LSI-MS(PI): MH+(amine): 286(60%), 287(100%), 288(45%) + F NH3 - O a 9 10 N b N 5 4 O N 8 7 6 1 F F O 2 NH 3 O 1H-NMR (400 MHz, DMSO-d6): = 2.40(3H, s, CH3 at C7), 2.50(3H, s, CH3 at C8), 3.23(2H, m, Hb ), 4.85(2H, t, J = 6.4 Hz, Ha), 7.80(1H, s, H9), 7.91(1H, s, H6), 7.99(3H, s, br, NH3+), 11.40(1H, s, NH). determination was done by HMBC and HSQC, first order coupling interpretation. 13C-NMR (100,6 MHz, DMSO-d6): = 18.6(CH3 at C7), 20.5(CH3 at C8), 36.4(Cb), 41.2(Ca), 115.6(C9), 117.1(q, JCF3-19F = 301.92 Hz, CF3), 130.4(C9a), 131.2(C6), 133.8(C5a), 135.9(C7), 137.2(C4a), 146.8(C8), 151.0(C10a), 155.4(C2), 158.0(q, JCOO(CF3)-19F = 31.33 Hz, COO), 159.7(C4). determination was done by HMBC and HSQC. N(10)-[4-(aminopropyl] flavin • TFA, CofC3 • TFA C17H18F3N5O4; 413.35 g/mol; LSI-MS(PI): MH+(amine): 300(4%), 301(7%), 302(5%) S5 + 9 10 N O N 1 8 6 -O b a 7 F NH3 c N 5 4 F F O 2 NH 3 O 1H-NMR (600 MHz, DMSO-d6): = 2.08(2H, m, Hb), 2.40(3H, s, CH3 at C7), 2.51(3H, s, CH3 at C8), 2.97(2H, m, Hc), 4.66(2H, t, J = 6.7 Hz, Ha), 7.83(1H, s, H9), 7.89(3H, s, br, NH3+), 7.92(1H, s, H6), 11.37(1H, s, NH). determination was done by HMBC and HSQC, first order coupling interpretation. 13C-NMR (150,9 MHz, DMSO-d6): = 18.7(CH3 at C7), 20.5(CH3 at C8), 25.0(Cb), 36.2(Cc), 41.4(Ca), 115.9(C9), 117.4(q, JCF3-19F = 299.31 Hz, CF3), 130.5(C9a), 131.1(C6), 133.9(C5a), 136.0(C7), 137.2(C4a), 146.7(C8), 150.5(C10a), 155.5(C2), 157.8(q, JCOO(CF3)-19F = 31.73 Hz, COO), 159.9(C4). determination was done by HMBC and HSQC. N(10)-[4-(aminopentyl] flavin • TFA, CofC5 • TFA C19H22F3N5O4; 441.41 g/mol; EI-MS(PI) : M+(amine): 327(30%), 328(7%) EI-MS(PI), high resolution: calculated: 327.1695, found: 327.1687, = 2.5 ppm. + NH3 e d c a N 8 6 N 5 O F F b 9 10 N 7 F -O 1 O 2 NH 3 4 O 1H-NMR (600 MHz, DMSO-d6): = 1.48(2H, m, Hc), 1.62(2H, m, Hd), 1.74(2H, m, Hb), 2.39(3H, s, CH3 at C7), 2.50(3H, s, CH3 at C8), 2.81(2H, m, He), 4.57(2H, m, Ha), 7.74(3H, s, br, NH3+), 7.77(1H, s, H9), 7.88(1H, s, H6), 11.31(1H, s, NH). determination was done by HMBC and HSQC. 13C-NMR (150,9 MHz, DMSO-d6): = 18.7(CH3 at C7), 20.6(CH3 at C8), 22.8(Cc), 25.9(Cb), 26.6(Cd), 38.6(Ce), 43.8(Ca), 116.0(C9), 117.2(q, JCF3-19F = 298.81 Hz, CF3), 130.7(C9a), 131.0(C6), 133.8(C5a), 135.8(C7), 137.1(C4a), 146.6(C8), 150.1(C10a), 155.7(C2), 158.0(q, JCOO(CF3)-19F = 31.18 Hz, COO), 159.9(C4). determination was done by HMBC and HSQC. N(10)-[4-(aminohexyl] flavin • TFA, CofC6 • TFA Synthesis is described in the supporting information of ref.1 C20H24F3N5O4; 455.43 g/mol S6 N(10)-[4-(aminooctyl] flavin • TFA, CofC8 • TFA C22H28F3N5O4; 483.49 g/mol; EI-MS(PI) : M+(amine): 369(60%), 370(10%) EI-MS(PI), high resolution: calculated: 369.2165, found: 369.2171, = 1.7 ppm. + NH 3 h g f e 9 6 10 N N 5 F b N 8 7 O F d c a F -O 1 O 2 NH 3 4 O 1H-NMR (600 MHz, DMSO-d6): = 1.32(4H, m, He, Hf), 1.36(2H, m, Hd), 1.47(2H, m, Hc), 1.53(2H, m, Hg), 1.72(2H, m, Hb), 2.40(3H, s, CH3 at C7), 2.52(3H, s, CH3 at C8), 2.78(2H, m, Hh), 4.57(2H, m, Ha), 7.67(3H, s, br, NH3+), 7.78(1H, s, H9), 7.91(1H, s, H6), 11.29(1H, s, NH). determination was done by HMBC, HSQC, COSY, and ROESY. 13C-NMR (150,9 MHz, DMSO-d6): = 18.7(CH3 at C7), 20.5(CH3 at C8), 25.6(Cf), 25.9(Cc), 26.3(Cb), 26.8(Cg), 28.2(Ce), 28.4(Cd), 38.7(Ch), 44.0(Ca), 115.9(C9), 117.2(q, JCF3-19F = 301.57 Hz, CF3), 130.6(C9a), 131.0(C6), 133.7(C5a), 135.7(C7), 137.0(C4a), 146.5(C8), 149.9(C10a), 155.6(C2), 157.7(q, JCOO(CF3)-19F = 30.26 Hz, COO), 159.9(C4). determination was done by HMBC, HSQC, COSY, and ROESY. Coupling of the flavins to DNA The protected DNA at the solid support (CPG) was washed with absolute MeCN and dried in vacuo. Thereafter, a solution of 30 – 50 mg of N,N’-Carbonyldiimidazol (CDI) in 1 ml DMSOabs. was added under argon atmosphere and mixed from time to time (syringes were applied). After 30-60 minutes the reaction mixture was removed and the CPG was washed 4 – 5 times with 10 ml DMSOabs. under argon atmosphere. In the next step 15-20 mg of the flavin (TFA-salt) was added in 1 ml DMSOabs. under argon atmosphere containing 3 – 5 drops NEt3 (in order to remove the TFA). The reaction was allowed to proceed for 2 days. Then the reaction mixture was removed and the CPG was washed first 3 times with 10 ml DMSOabs., thereafter with 10 ml MeCNabs., and dried in vacuo. Cleavage and deprotection of the DNA was carried out in 700 µl aqueous ammonia solution (30 %) for 48 h at 30 °C. The solid support was filtered off and washed twice with 300 µl H2O. The ammonia was removed from the combined aqueous solution with a speedvac and the solid DNA was obtained after lyophilization. Further purification was carried out by HPLC (MeCN/H2O/NH4+CH3COO-, 50 mM). The purity of the resulting oligomer was controlled by analytical HPLC, characterization was done by mass spectroscopy. Representatively, the MS signals of some flavin-DNA ligands are listed. S7 CofC2-O5: C63H75N23O31P4; 1774.32 g/mol; ESI-MS(PI) : MH+: 1774.6, 1775.0, 1775.9, 1776.6, 1777.0; M+2H+: 887.9; M+3H+: 592.3; ESI-MS(NI): M-H+: 1773.0, 1773.9, 1774.9; M-2H+: 885.9, 886.4; M-3H+: 590.3. CofC4-O5: C65H79N23O31P4; 1802.37 g/mol; ESI-MS(PI) : MH+: 1803.1, 1804.3, 1804.8; M+2H+: 901.9; M+3H+: 601.7; ESI-MS(NI): M-H+: 1801.0, 1801.7, 1802.1, 1802.7; M-2H+: 900.0, 900.4; M3H+: 599.6. CofC6-O5: C67H83N23O31P4; 1830.43 g/mol; ESI-MS(PI): MH+: 1831.3: M+2H+: 915.9, 916.4; M+3H+: 611.1; ESI-MS(NI): M-H+: 1828.0, 1828.4, 1829.1, 1829.9, 1831.2; M-2H+: 914.0, 914.5 CofC4-O20: C212H263N80O122P19; 6472.39 g/mol; ESI-MS(PI): M+2H++Na+: 2165.6; M+3H++Na+: 1624.6; M+4H+: 1619.2; ESI-MS(NI): M-4H++Na+: 2164; M-3H+: 2156.6, 2157.4; M-5H++Na+: 1623.1; M-4H+: 1617.1; M-6H++Na+: 1298.1; M-5H+: 1293.4. CofC6-O20: C214H267N80O122P19; 6500.44 g/mol; ESI-MS(PI): M+3H+: 2167.5; M+4H+: 1626.1, 1626.5; ESI-MS(NI): M-3H+: 2165.9; M-4H+: 1624.1. S8 Combined SPR and electrochemical measurements An SPR-setup according to ref.4 was applied in combination with an EG&G Potentiostat/Galvanostat Model 273 or with an Amel Potentiostat/Galvanostat 2053. The gold surface (working electrode) was contacted by a Pt foil, another Pt foil served as pseudo-reference and a gold coated wire as counter electrode. Generation of dodecin variants Introduction of point mutation is described elsewhere.5 The Glu50, Glu51 deletion was introduced by the megaprimer method,6 using primers: MP-MUT1rev (pDOD-delEE): CCGTCTGATACGTGCGGACGGCGCCGATTTCGACGCCCTGG MP-MUT2for (pDOD-delEE): CGAAATCGGCGCCGTCCGCACGTATCAGACGGAAGTGCAGG X-ray structural analysis Crystallization of dodecin is described elsewhere.5 Data on dodecin holocomplex crystals were collected to 1.8 Å (apododecin/CofC4-O5) and 1.7 Å (apododecin/ CofC2-O5). Diffraction intensities were integrated with XDS and scaled with XSCALE7. Initial models (template 2ccc) were refined by cycles of model building in COOT8 and automatic refinement in REFMAC9-12. Figures of crystal structures were created with PYMOL (http://pymol.org). Table S1. Data Collection and Refinement Statistics data set CofC4-O5 holocomplex CofC2-O5 holocomplex Data Collection Statistics pdb accession code X-ray source/ detector system 2vkg 2vkf ESRF,BM14/ SLS,PXII/ mar225 mosaic CCD mar225 mosaic CCD S9 Wavelength [Å] 0.976 1.033 Cell constants [Å] a = b = c = 142.03 a = b = c = 142.70 Resolution range [Å] 20.0–1.80 (1.90–1.80) 20.0–1.69 (1.80–1.69) Observations 107589 (16720) 171130 (14542) unique reflections 11640 (1808 ) 13434 (1732) 22.94 21.66 [%] 10.2 (58.5) 9.3 (51.9) Completeness [%] 97.1 (96.7) 95.4 (79.1) I/σ(I) 19.18 (3.44) 19.8 (4.1) Wilson [Å2] aR merge B-factor Refinement Statistics (REFMAC) Space group F4132 F4132 19.75-1.80 29-1.7 (1.847-1.80) (1.744-1.70) 11077 (796) 12770 (730) 19.2/21.3 (26.9/34.8) 18.4/21.3 (26.7/32.4) Atoms 605 621 Water molecules 86 86 0.011 0.019 1.24 1.42 19.364 22.8 Most favored [%] 91.2 93 Additional allowed [%] 8.8 7 Generously allowed [%] 0 0 Disallowed [%] 0 0 Resolution range [Å] Unique reflections bR c cryst/ Rfree [%] Geometry Rmsd of length [Å] bond Rmsd of bond angles [degree] Mean [Å2] B value Ramachandran Rmerge = Σ | I - <I>| / ΣI, where I is the observed intensity and <I> the average intensity from multiple observations of symmetry-related reflections; values in parentheses correspond to the highest resolution shell b Rcryst = Σ | (Fobs) - (Fcalc) | / Σ (Fobs) c Rfree = crystallographic R-factor based on 5% of the data withheld from the refinement for cross-validation a S10 SPR-Measurements SPR was recorded through the thin metal film in the so-called Kretschmann configuration.13 Gold films of typically 40 to 50 nm were vacuum evaporated on glass slides (LaSFn9, n = 1.844 at 633 nm). The gold-coated glass slides were used for immobilization experiments without further treatment. In the SPR setup, they were optically coupled to the base of a 90° prism of the same material by index-matching. For electrochemical experiments, the SPR cuvette was equipped with 3 electrodes; the gold coated slide serving as a working electrode, a gold-coated counter electrode and a Pt pseudoreference electrode. The energy coupling in the surface plasmon could be observed as a deep minimum in the reflectivity of the p-polarized light when the angle of incidence in the SPR setup was incremented. Reflectivity vs. angle of incidence scans yielded the optical thickness of the adsorbed coating. Immobilization kinetics were recorded by monitoring the reflected light intensity at a fixed angle as a function of time. From the Fresnel fit to the resonance curve for a bare gold surface, the dielectric constant and thickness of the gold layer can be obtained. Adding layers to the surface of the gold, as obtained in respective coating steps, generally shifts scan curves towards higher angles. Fitting these curves determines the optical thickness of the layers.14,15 For DNA as well as for the protein a refractive index n of 1.375 was assumed.16 SPR angle shifts were calculated using the Fresnel equations via SPR software (Winspall version 3.01, Max Planck Institute of Polymer Research, Mainz, Germany). Preparation of ds-DNA monolayers (CofC4-ds20-S-Au) Direct Adsorption Protocol: CofC4-O20, and RS-O20 were hybridized in PBS buffer (adjusted to 1 M NaCl) by heating an equimolar solution of these oligonucleotides to 75 °C for 5 minutes and slowly cooling to ambient temperatures. The generated ds-DNA (CofC4-ds20-SR) was injected into the fluid cell of the SPR system at a concentration of 100 µM. After incubation for 2 hours the gold surface was washed with buffer (PBS buffer adjusted to 1 M NaCl), until a stable baseline in reflectivity was reached. Before applying the tE variant, the immobilization experiments were routinely started with the dA non-binding variant as negative control. S11 Tarlov Protocol: In contrast to the direct adsorption approach, in the Tarlov protocol the ds-DNA hybridization was performed on the gold surface. First the ss-DNA RS-O20, or alternatively the free thiol derivative HSO20, both at concentrations 8 µM in 100 mM Tris-HCl pH 7.5 was adsorbed on gold (for deprotection of RS-O20 with tris(2-carboxyethyl) phosphine (TCEP) to yield HS-O20 see ref.17). The incubation period was varied in different experiments between 20 and 90 min. Next, the ss-DNA coated surface was treated with mercaptohexanol (1 mM) for a period of 5 to 10 min in order to release nonspecifically adsorbed ss-DNA and to orient the specifically adsorbed DNA. In the following step, the fluid cell was equilibrated to 1 M NaCl, 10 mM Tris-HCl pH 7.5, 1 mM EDTA, before hybridizing with a 5 µM solution of CofC4-O20 during an incubation period of about 2 hours. The surface was rinsed and finally equilibrated to protein binding buffer (PBS buffer adjusted to 1 M NaCl) for dodecin immobilization experiments. Similarly to the direct adsorption protocol, the immobilization experiments were routinely started with the dA non-binding variant. In the direct adsorption as well as the Tarlov protocol, the modified surface was kept immersed in buffer solution throughout the entire procedure. The (apo)proteins dA and tE were used in concentrations of 20 µM (in PBS adjusted to 1 M NaCl) with 0.5 equivalents (10 µM) of lumichrome as a binding mediator. For details of the dA and tE protein immobilizations see legends of Figure 5 and 8 in the main manuscript. S12 Re-Binding of tE R [%] d1 d2 42 42 40 40 38 38 0 10 20 30 0 3 6 time [min] Figure S1. Binding and re-binding after reductive release of the variant tE on CofC4-ds-S-Au layers generated by the direct adsorption protocol. (A) d1) Dodecin tE was adsorbed on CofC4-ds-S-Au monolayer in PBS buffer adjusted to 1 M NaCl (end of incubation is omitted in this Figure). d2) After the release of the protein by applying a negative potential to -600 mV (vs. the Pt pseudoreference electrode), the tE variant re-binds at zero potential. The gain in reflectivity (surface layer thickness) is about 40 % of the preceding decrease during reduction. The dashed lines highlight the start of incubation with the binding variant tE. S13 SPR Angular Scan Curves Collected for an Experiment Following the Tarlov Procedure 80 1 2 3 4 15 40 20 Ref-Koeff R [%] Ref-Koeff R [%] 60 5 10 1 6 2 6 5 3 4 5 57 58 59 60 61 Theta [°] 0 45 50 55 60 Theta [°] Figure S2. Angular scan curves of the reflectivity at various steps of an experiment following the Tarlov procedure. Different angular scan curves are labeled as follows: (1) bare gold surface in 100 mM Tris-HCl (pH 7.5), (2) ss-DNA monolayer in 100 mM Tris-HCl (pH 7.5) (generated by adsorption of HS-O20), (3) ssDNA monolayer in 1 M NaCl, 10 mM Tris-HCl (pH 7.5), 1 mM EDTA, (4) CofC4-ds20-S-Au monolayer in 1 M NaCl, 10 mM Tris-HCl (pH 7.5), 1 mM EDTA, (5) CofC4-ds20-S-Au monolayer in PBS adjusted to 1 M NaCl and (6) tE bound to CofC4-ds20-S-Au monolayer in PBS adjusted to 1 M NaCl. The SPR angles, at which the intensity is minimal at the curves, shift during the process of selfassembly of substrates and were fitted to surface layer thicknesses. After assembly the steps, the goldcoated substrates were equilibrated to the buffer conditions of the next immobilization step. Accordingly, immobilization of HS-O20 was fitted to an increase in surface layer thickness of 3.0 nm. Self assembly to CofC4-ds20-S-Au by adding CofC4-O20 further increased layer thickness by 0.9 nm (3.9 nm overall surface layer thickness of CofC4-ds20-S-Au). Rinsing the CofC4-ds20-S-Au layer with the dodecin variant tE led to specific binding of protein and increased the surface layer thickness by 6.3 nm (scan curve of the non-binding dA variant not shown). The diameter of dodecin is 7 nm and, thus, the angular scan curves suggest a tightly packed monolayer of dodecin bound to the flavin moiety of CofC4-ds20-S-Au. The inset shows a zoom-in to the angles between 56 and 61°. S14 Kinetics of Protein Immobilization Kinetics of protein immobilizations were fitted to a Langmuir Isotherm, p = R0+(Rmax-R0)*(1-e(-(k)*(tdt))), where k = kon*c+koff. R0 is the initial and Rmax the maximum reflectivity, dt the start of kinetics and c the protein concentration. In the respective fit function the protein concentration c was set 1/12 of the molar concentration of monomer/binding positions, as just one of the 12 binding positions is occupied by the immobilized ligand. In binding experiments in solution, we observed an increase in affinity of CoFCn-O5 ligands when lumichrome was present. As we expected lumichrome to saturate semioccupied binding pockets by binding to remaining binding positions, lumichrome was added to the buffer in protein immobilization steps (see above). A B 63 54 62 61 52 Model: langmuir Chi^2/DoF = 0.00031 R^2 = 0.99955 Ro 50.68978 ±0.00518 Rmax 53.71421 ±0.00527 k 0.00414 ±0.00002 51 50 0 500 1000 1500 2000 R [%] R [%] 53 60 Model: langmuir Chi^2/DoF = 0.00184 R^2 = 0.99915 Ro 57.45519 ±0.02295 Rmax 62.93785 ±0.01308 k 0.01365 ±0.00014 59 58 57 0 2500 150 300 450 600 time [sec] time [sec] C R [%] 34 32 Model: langmuir Chi^2/DoF = 0.07235 R^2 = 0.9394 Ro 30 ±0 Rmax 34 ±0 k 0.00344 ±0.00012 30 0 500 1000 1500 time [sec] 2000 2500 Figure S3. Kinetics of tE dodecin binding fitted to a Langmuir Isotherm. The gray dashed line displays the start of incubating the ds-DNA coated surface (CofC4-ds20-S-Au) with the dodecin variant tE. The arrow indicated rinsing the surface with protein-free solution (not shown in C). As koff values were assumed to be very low, kon values were calculated according to kon = k/c to 2.48 · 103 M-1s-1 (A), 8.2 · 103 M-1s-1 (B), and 2.06 · 103 M-1s-1 (C). (A and B) Surface layer along Tarlov procedure; (C) along direct adsorption procedure: At the beginning of protein immobilization, the increase of S15 reflectivity could be fitted to the Langmuir Isotherm. When the signal asymptotically reaches its final value, deviations from the fit functions were observed. This might be caused by steric interactions of the monomers. At the start and the end of tE incubation, we frequently observed changes in reflectivity due to a change in the refractive indices of buffers used. This is seen especially in Figure S3A, where at the start of incubation (gray dashed line) the read-out signal drops, but is regained when re-equilibrated to lumichrome-free (protein-free) buffer (arrow). References (1) Nöll, G.; Kozma, E.; Grandori, R.; Carey, J.; Schoedl, T.; Hauska, G.; Daub, J. Langmuir 2006, 22, 2378. (2) Epple, R.; Carell, T. J. Am. Chem. Soc. 1999, 121, 7318. (3) Epple, R.; Wallenborn, E.-U.; Carell, T. J. Am. Chem. Soc. 1997, 119, 7440. (4) Ekgasit, S.; Thammacharoen, C.; Yu, F.; Knoll, W. Anal. Chem. 2004, 76, 2210. (5) Grininger, M.; Zeth, K.; Oesterhelt, D. J. Mol. Biol. 2006, 357, 842. (6) Sarkar, G.; Sommer, S. S. BioTechniques 1990, 8, 404. (7) Kabsch, W. J. Appl. Crystallogr. 1988, 21, 67. (8) Emsley, P.; Cowtan, K. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2004, D60, 2126. (9) Brunger, A. T.; Adams, P. D.; Clore, G. M.; DeLano, W. L.; Gros, P.; Grosse-Kunstleve, R. W.; Jiang, J.-S.; Kuszewski, J.; Nilges, M.; Pannu, N. S.; Read, R. J.; Rice, L. M.; Simonson, T.; Warren, G. L. Acta Crystallogr., Sect. D: Biol. Crystallogr. 1998, D54, 905. (10) Jones, T. A.; Zou, J. Y.; Cowan, S. W.; Kjeldgaard, M. Acta Crystallogr., Sect. A: Found. Crystallogr. 1991, A47, 110. (11) Murshudov, G. N.; Vagin, A. A.; Dodson, E. J. Acta Crystallogr., Sect. D: Biol. Crystallogr. 1997, D53, 240. (12) Meissner, B.; Schleicher, E.; Weber, S.; Essen, L.-O. J. Biol. Chem. 2007, 282, 33142. (13) Kretschmann, E. Z. Phys. 1971, 241, 313. (14) Knoll, W. Annu. Rev. Phys. Chem. 1998, 49, 569. (15) Raether, H. Surface plasmons on smooth and rough surfaces and on gratings; SpringerVerlag, 1988; Vol. 111. (16) Liu, J.; Tiefenauer, L.; Tian, S.; Nielsen, P. E.; Knoll, W. Anal. Chem. 2006, 78, 470. (17) Nogues, C.; Cohen, S. R.; Daube, S. S.; Naaman, R. Phys. Chem. Chem. Phys. 2004, 6, 4459. S16