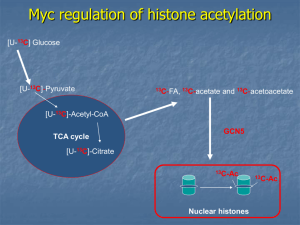
HEP_26302_sm_SuppFig
advertisement

Supporting Figures Supporting Fig. 1. Myc is overexpressed in human liver tumor cells. Western Blot assays showing Myc protein levels in indicated cell lines. 1 Supporting Fig. 2. Myc is essential for malignant phenotypes in HepG2 and BEL-7402 cell lines. (A) Western Blot assays showing change of Myc protein levels in Myc-siRNA cells. (B) Focus formation assays of HepG2 or BEL-7402 cell lines harboring Myc siRNA. (C) Soft-agar formation assays of HepG2 or BEL-7402 cell lines with Myc siRNA transfection. Down panel: Quantification of soft agar colony numbers of the soft agar experiments. (D) Cell cycle analyses. Cells were harvested in log-phase growth. Nuclei were stained with propidium iodide and analyzed by flow cytometry. (E) Expression of Myc in HepG2 or BEL-7402 with differential concentration AF10058-F4 treatment. (F) Focus formation assay of HepG2 or BEL-7402 cells with AF10058-F4 treatment as indicated concentration. 2 Supporting Fig. 3. Induction of Myc increases malignant phenotypes in HL7702 cells. (A) Real-time RT-PCR showing change of Myc mRNA (normalized to GADPH) in HL-7702-MycER cells with induction of Myc. (B) Anchorage-dependent soft agar growth of HL-7702-MycER with induction of Myc. 3 Supporting Fig. 4. Large scale pre-screening for promoter E-boxes in CpG islands from 1049 miRNA genomic loci. (A) Summary of pre-screening results. Among 1049 miRNA genomic loci, 834 loci scored positive with one or more E-boxes within the searching boundaries. E-boxes from 94 loci are located in CpG islands among above 834 loci. E-boxes from 21 loci are conserved between human and mouse among above 94 loci. (B)Venn diagram of distribution of 21 miRNA loci. miRNAs from 10 loci have been reported to be directly regulated by Myc among above 21 loci. miRNAs from the other 11 loci have not been reported to be regulated by Myc (C) Chromosome location of above 21 miRNA loci. 4 Supporting Fig. 5. Myc regulates other miRNA expression. (A) Schematic representation of genomic structures of indicated miRNAs. (B) Quantitative PCR CHIP analysis of Myc binding to differential genomic region in BEL7402 cells. (C) Conventional PCR and agarose-gel analyses. 5 Supporting Fig. 6. Other miRNAs inhibit colony formation. (A) Other miRNA expression levels normalized for U6 by qRT-PCR. (B) Foci formation analyses of indicated cells. (C) Soft agar colony formation assay of indicated cells. 6 Supporting Fig. 7. Ectopic expression of miR-148a-5p or miR-363-3p inhibits migration of HepG2 and BEL-7402 cells. Indicated cells were cultured to 90% confluence in six-well plates. A wound was made by dragging an autoclaved plastic pipette tip across the cell surface and cells were maintained in serum-free medium. The phase contrast images of the wounds were recorded for incubations of 0, 48 and 72 hours, and all experiments were triplicated. 7 Supporting Fig. 8. miR-148a-5p directly targets Myc in 293T cells. (A) GFP reporter analyses in 293T cells. GFP reporter construct containing Myc UTR was co-trasfected with control or miR-148a-5p mimics. GFP photographs were taken 48hr later after transfection. (B) Relative percent of GFP of indicated samples. (C) Myc mRNA level normalized for GAPDH by qRT-PCR. (D) Myc protein level after transfected with miR-148a-5p. 8 Supporting Fig. 9. USP28 is a direct target of miR-363-3p in 293T cells. (A) GFP reporter analyses in 293T cells. The experimental procedure is as shown in supplemental Fig. 8. (B) Relative percent of GFP of indicated samples. (C) USP28 and Myc mRNA levels normalized for GAPDH by qRT-PCR. (D) USP28 protein level after transfected with miR-363-3p. 9 Supporting Fig. 10. Inhibition of miR-148a-5p or miR-363-3p promotes migration of HL7702 and FHCC98 cells. The experimental procedure is as shown in supplemental Fig. 7. 10