EXERCISE 3 MULIPLE ALIGNMENT AND BLAST
advertisement

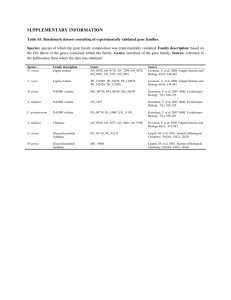
EXERCISE 3 MULIPLE ALIGNMENT AND BLAST Suppose you have cloned and sequenced the following sequence in the lab. During the exercise session we will try to study the function of this gene based on in silico searches. The only thing we know is that we isolated this sequence from the bacterial species Paracoccus denitrificans and that it is involved in respiration. MADAAVHGHGDHHDTRGFFTRWFMSTNHKDIGILYLFTAGIVGLISVCFTVYMRMELQHPGVQYMCLEGARLIADASAEC TPNGHLWNVMITYHGVLMMFFVVIPALFGGFGNYFMPLHIGAPDMAFPRLNNLSYWMYVCGVALGVASLLAPGGNDQMGS GVGWVLYPPLSTTEAGYSMDLAIFAVHVSGASSILGAINIITTFLNMRAPGMTLFKVPLFAWSVFITAWLILLSLPVLAG AITMLLMDRNFGTQFFDPAGGGDPVLYQHILWFFGHPEVYIIILPGFGIISHVISTFAKKPIFGYLPMVLAMAAIGILGF VVWAHHMYTAGMSLTQQAYFMLATMTIAVPTGIKVFSWIATMWGGSIEFKTPMLWAFGFLFLFTVGGVTGVVLSQAPLDR VYHDTYYVVAHFHYVMSLGAVFGIFAGVYYWIGKMSGRQYPEWAGQLHFWMMFIGSNLIFFPQHFLGRQGMPRRYIDYPV EFAYWNNISSIGAYISFASFLFFIGIVFYTLFAGKRVNVPNYWNEHADTLEWTLPSPPPEHTFETLPKREDWDRAHAH 1 convert file to FastA format The FastA format is the standard format used by most sequence based programs (clustalW, …) A sequence in FASTA format begins with a single-line description, followed by lines of sequence data. The description line is distinguished from the sequence data by a greater-than (">") symbol in the first column. It is recommended that all lines of text be shorter than 80 characters in length. An example sequence in FASTA format is: >gi|532319|pir|TVFV2E|TVFV2E envelope protein ELRLRYCAPAGFALLKCNDADYDGFKTNCSNVSVVHCTNLMNTTVTTGLLLNGSYSENRTQIWQKHRTSNDS ALILLNKHYNLTVTCKRPGNKTVLPVTIMAGLVFHSQKYNLRLRQAWCHFPSNWKGAWKEVKEEIVNLPKER YRGTNDPKRIFFQRQWGDPETANLWFNCHGEFFYCKMDWFLNYLNNLTVDADHNECKNTSGTKSGNKRAPG PCVQRTYVACHIRSVIIWLETISKKTYAPPREGHLECTSTVTGMTVELNYIPKNRTNVTLSPQIESIWAAELDRY KLVEITPIGFAPTEVRRYTGGHERQKRVPFVXXXXXXXXXXXXXXXXXXXXXXVQSQHLLAGILQQQKNLL AAVEAQQQMLKLTIWGVK Sequences are expected to be represented in the standard IUB/IUPAC amino acid and nucleic acid codes, with these exceptions: lower-case letters are accepted and are mapped into upper-case; a single hyphen or dash can be used to represent a gap of indeterminate length; and in amino acid sequences, U and * are acceptable letters (see below). Before submitting a request, any numerical digits in the query sequence should either be removed or replaced by appropriate letter codes (e.g., N for unknown nucleic acid residue or X for unknown amino acid residue). The nucleic acid codes supported are: A --> adenosine M --> A C (amino) C --> cytidine S --> G C (strong) G --> guanine W --> A T (weak) T --> thymidine B --> G T C U --> uridine D --> G A T R --> G A (purine) H --> A C T Y --> T C (pyrimidine) V --> G C A K --> G T (keto) N --> A G C T (any) - gap of indeterminate length For those programs that use amino acid query sequences (BLASTP and TBLASTN), the accepted amino acid codes are: A alanine P proline B aspartate or asparagine Q glutamine C cystine R arginine D aspartate S serine E glutamate T threonine F phenylalanine U selenocysteine G glycine V valine H histidine W tryptophan I isoleucine Y tyrosine K lysine Z glutamate or glutamine L leucine X any M methionine * translation stop N asparagine - gap of indeterminate length This is the sequence of the unknown gene > unknown gene MADAAVHGHGDHHDTRGFFTRWFMSTNHKDIGILYLFTAGIVGLISVCFTVYMRMELQHPGVQYMCLEGAR LIADASAECTPNGHLWNVMITYHGVLMMFFVVIPALFGGFGNYFMPLHIGAPDMAFPRLNNLSYWMYVCGV ALGVASLLAPGGNDQMGSGVGWVLYPPLSTTEAGYSMDLAIFAVHVSGASSILGAINIITTFLNMRAPGMTLF KVPLFAWSVFITAWLILLSLPVLAGAITMLLMDRNFGTQFFDPAGGGDPVLYQHILWFFGHPEVYIIILPGFGIIS HVISTFAKKPIFGYLPMVLAMAAIGILGFVVWAHHMYTAGMSLTQQAYFMLATMTIAVPTGIKVFSWIATMW GGSIEFKTPMLWAFGFLFLFTVGGVTGVVLSQAPLDRVYHDTYYVVAHFHYVMSLGAVFGIFAGVYYWIGK MSGRQYPEWAGQLHFWMMFIGSNLIFFPQHFLGRQGMPRRYIDYPVEFAYWNNISSIGAYISFASFLFFIGIVFY TLFAGKRVNVPNYWNEHADTLEWTLPSPPPEHTFETLPKREDWDRAHAH Homology search using Blast http://www.ncbi.nlm.nih.gov/guide/homology/ Find homologs with your sequence of interest in the NCBI sequence database. Blast your sequence. Can you derive from the blasthits a clue on the function of your sequenced protein? Go to the GenBank file of the best hit: (click on the link) What information can you find in the GenBank file? Do you find Matches with eukaryotic sequences? Are these Significant? What does that mean. This system is interesting because it is very ancient and has been conserved throughout all phylogenetic branches. To find out which sequence residues are involved in the catalytic function, we will construct an alignment of sequences from distinct species, so that we can have a representative alignment of the family of terminal oxidases. From this alignment we will derive the residues that are essential for the function because they have been conserved. (perform a query against the reference database) In what species do find most hits? Is this what you expect why? Select from this file the following sequences in FastA format and add them to the clipboard Select 3 sequences from these bacterial species: eg Items 1 - 3 of 3 One page. 1. cytochrome-c oxidase [Paracoccus denitrificans PD1222] 558 aa protein YP_915727.1 GI:119384671 2. cytochrome c oxidase, aa3-type, subunit I [Rhodobacterales bacterium HTCC2654] 557 aa protein ZP_01014069.1 GI:84686174 3. putative cytochrome C oxidase polypeptide I transmembrane protein [Sinorhizobium meliloti 1021] 562 aa protein NP_385011.1 GI:15964658 Because the protein is so widely distributed and we found some hits in eukaryotes as well, we will search for more eukaryotic hits. However, because the “unknown protein” is from bacterial origin we will retrieve all prokaryotic sequences first because they will be most similar. To focus on the eukaryotic hits, we will perform an advanced blastsearch and blast the “unknown sequence” against the non redundant database from which the prokaryotic sequences were excluded. Redo the blast but now only search for mammalian hits Redo the blast but know only search for mammalian hits Add a few accession numbers but definitely select from the list Add from the selected sequences from the blast output to the clipboard 1: ABR93038 Reports BLink, Conserved Domains, Links cytochrome c oxidase subunit I [Homo sapiens] gi|151327759|gb|ABR93038.1|[151327759] 2: YP_001686700 Reports BLink, Conserved Domains, Links cytochrome c oxidase subunit I [Mus musculus musculus] gi|167716839|ref|YP_001686700.1|[167716839] Redo the blast but now only search for plant hits viridaeplantae 3: NP_085587 Reports BLink, Conserved Domains, Links cytochrome c oxidase subunit 1 [Arabidopsis thaliana] gi|13449404|ref|NP_085587.1|[13449404] 4: YP_514675 Reports BLink, Conserved Domains, Links cytochrome c oxidase subunit 1 [Oryza sativa Indica Group] gi|89280750|ref|YP_514675.1|[89280750] Add the following sequences by searching for the following accessionnumbers in GenBank, Add the FastA files to the clipboard. YP_428413 NP_769403 YP_001188087 Download the corresponding protein sequences in FastA format >gi|151327759|gb|ABR93038.1| cytochrome c oxidase subunit I [Homo sapiens] MFADRWLFSTNHKDIGTLYLLFGAWAGVLGTALSLLIRAELGQPGNLLGNDHIYNVIVTAHAFVMIFFMV MPIMIGGFGNWLVPLMIGAPDMAFPRMNNMSFWLLPPSLLLLLASAMVEAGAGTGWTVYPPLAGNYSHPG ASVDLTIFSLHLAGVSSILGAINFITTIINMKPPAMTQYQTPLFVWSVLITAVLLILSLPVLAAGITMLL TDRNLNTTFFDPAGGGDPILYQHLFWFFGHPEVYILILPGFGMISHIVTYYSGKKEPFGYMGMVWAMMSI GFLGFIVWAHHMFTVGMDVDTRAYFTSATMIIAIPTGVKVFSWLATLHGSNMKWSAAVLWALGFIFLFTV GGLTGIVLANSSLDIVLHDTYYVVAHFHYVLSMGAVFAIMGGFIHWFPLFSGYTLDQTYAKIHFTIMFIG VNLTFFPQHFLGLSGMPRRYSDYPDAYTTWNILSSVGSFISLTAVMLMIFMIWEAFASKRKVLMVEEPSM NLEWLYGCPPPYHTFEEPVYMKS >gi|167716839|ref|YP_001686700.1| cytochrome c oxidase subunit I [Mus musculus musculus] MFINRWLFSTNHKDIGTLYLLFGAWAGMVGTALSILIRAELGQPGTLLGDDQIYNVIVTAHAFVMIFFMV MPMMIGGFGNWLVPLMIGAPDMAFPRMNNMSFWLLPPSFLLLLASSMVEAGAGTGWTVYPPLAGNLAHAG ASVDLTIFSLHLAGVSSILGAINFITTIINMKPPAMTQYQTPLFVWSVLITAVLLLLSLPVLAAGITMLL TDRNLNTTFFDPAGGGDPILYQHLFWFFGHPEVYILILPGFGIISHVVTYYSGKKEPFGYMGMVWAMMSI GFLGFIVWAHHMFTVGLDVDTRAYFTSATMIIAIPTGVKVFSWLATLHGGNIKWSPAMLWALGFIFLFTV GGLTGIVLSNSSLDIVLHDTYYVVAHFHYVLSMGAVFAIMAGFVHWFPLFSGFTLDDTWAKAHFAIMFVG VNMTFFPQHFLGLSGMPRRYSDYPDAYTTWNTVSSMGSFISLTAVLIMIFMIWEAFASKREVMSVSYAST NLEWLHGCPPPYHTFEEPTYVKVK >gi|13449404|ref|NP_085587.1| cytochrome c oxidase subunit 1 [Arabidopsis thaliana] MKNLVRWLFSTNHKDIGTLYFIFGAIAGVMGTCFSVLIRMELARPGDQILGGNHQLYNVLITAHAFLMIF FMVMPAMIGGFGNWFVPILIGAPDMAFPRLNNISFWLLPPSLLLLLSSALVEVGSGTGWTVYPPLSGITS HSGGAVDLAIFSLHLSGVSSILGSINFITTIFNMRGPGMTMHRLPLFVWSVLVTAFLLLLSLPVLAGAIT MLLTDRNFNTTFFDPAGGGDPILYQHLFWFFGHPEVYILILPGFGIISHIVSTFSGKPVFGYLGMVYAMI SIGVLGFLVWAHHMFTVGLDVDTRAYFTAATMIIAVPTGIKIFSWIATMWGGSIQYKTPMLFAVGFIFLF TIGGLTGIVLANSGLDIALHDTYYVVAHFHYVLSMGAVFALFAGFYYWVGKIFGRTYPETLGQIHFWITF FGVNLTFFPMHFLGLSGMPRRIPDYPDAYAGWNALSSFGSYISVVGICCFFVVVTITLSSGNNKRCAPSP WALELNSTTLEWMVQSPPAFHTFGELPAIKETKSYVK >gi|89280750|ref|YP_514675.1| cytochrome c oxidase subunit 1 [Oryza sativa Indica Group] MTNLVRWLFSTNHKDIGTLYFIFGAIAGVMGTCFSVLIRMELARPGDQILGGNHQLYNVLITAHAFLMIF FMVMPAMIGGFGNWFVPILIGAPDMAFPRLNNISFWLLPPSLLLLLSSALVEVGSGTGWTVYPPLSGITS HSGGAVDLAIFSLHLSGVSSILGSINFITTIFNMRGPGMTMHRLPLFVWSVLVTAFLLLLSLPVLAGAIT MLLTDRNFNTTFFDPAGGGDPILYQHLFWFFGHPEVYILILPGFGIISHIVSTFSRKPVFGYLGMVYAMI SIGVLGFLVWAHHMFTVGLDVDTRAYFTAATMIIAVPTGIKIFSWIATMWGGSIQYKTPMLFAVGFIFLF TIGGLTGIVLANSGLDIALHDTYYVVAHFHYVLSMGAVFALFAGFYYWVGKIFGRTYPETLGQIHFWITF FGVNLTFFPMHFLGLSGMPRRIPDYPDAYAGWNALSSFGSYISVVGIRRFFVVVAITSSSGKNKRCAESP WAVEQNPTTLEWLVQSPPAFHTFGELPAIKETKS >gi|69934593|ref|ZP_00629671.1| Cytochrome-c oxidase [Paracoccus denitrificans PD1222] MADAAVHGHGDHHDTRGFFTRWFMSTNHKDIGILYLFTAGIVGLISVCFTVYMRMELQHPGVQYMCLEGA RLIADASAECTPNGHLWNVMITYHGVLMMFFVVIPALFGGFGNYFMPLHIGAPDMAFPRLNNLSYWMYVC GVALGVASLLAPGGNDQMGSGVGWVLYPPLSTTEAGYSMDLAIFAVHVSGASSILGAINIITTFLNMRAP GMTLFKVPLFAWSVFITAWLILLSLPVLAGAITMLLMDRNFGTQFFDPAGGGDPVLYQHILWFFGHPEVY IIILPGFGIISHVISTFAKKPIFGYLPMVLAMAAIGILGFVVWAHHMYTAGMSLTQQAYFMLATMTIAVP TGIKVFSWIATMWGGSIEFKTPMLWAFGFLFLFTVGGVTGVVLSQAPLDRVYHDTYYVVAHFHYVMSLGA VFGIFAGVYYWIGKMSGRQYPEWAGQLHFWMMFIGSNLIFFPQHFLGRQGMPRRYIDYPVEFAYWNNISS IGAYISFASFLFFIGIVFYTLFAGKRVNVPNYWNEHADTLEWTLPSPPPEHTFETLPKREDWDRAHAH >gi|146276716|ref|YP_001166875.1| Cytochrome-c oxidase [Rhodobacter sphaeroides ATCC 17025] MADAAIHGHEHDRRGFFTRWFMSTNHKDIGVLYLFTGGLVGLISVAFTVYMRMELMAPGVQFMCAEHLES GLVKGFFQSLWPSAVENCTPNGHLWNVMITAHGILMMFFVVIPALFGGFGNYFMPLHIGAPDMAFPRMNN LSFWLYVAGTSLAVASLFAPGGNGQLGSGIGWVLYPPLSTSESGYSTDLAIFAVHLSGASSILGAINMIT TFLNMRAPGMTMHKVPLFAWSIFVTAWLILLALPVLAGAITMLLTDRNFGTTFFQPSGGGDPVLYQHILW FFGHPEVYIIVLPAFGIVSHVIATFSKKPIFGYLPMVYAMVAIGVLGFVVWAHHMYTAGLSLTQQSYFMM ATMVIAVPTGIKIFSWIATMWGGSIELKTPMLWALGFLFLFTVGGVTGIVLSQASVDRYYHDTYYVVAHF HYVMSLGAVFGIFAGIYFWIGKMSGRQYPEWAGKLHFWMMFVGANLTFFPQHFLGRQGMPRRYIDYPEAF ATWNFVSSLGAFLSFASFLFFIGIVFYTLTRGARVTANNYWNEHADTLEWTLTSPPPEHTFERLPKREDW DRSHAH >gi|27376282|ref|NP_767811.1| cytochrome C oxidase subunit I [Bradyrhizobium japonicum USDA 110] MATSAAAHGDHAQDHGHDEHAHPTGWRRYVYSTNHKDIGTMYLIFAVIAGVIGAAMSIAIRAELMYPGVQ IFHETHTYNVFVTSHGLIMIFFMVMPAMIGGFGNWFVPLMIGAPDMAFPRMNNISFWLLPASFGLLLMST FVEGEPGANGVGAGWTMYVPLSSSGHPGPAVDFAILSLHLAGASSILGAINFITTIFNMRAPGMTLHKMP LFVWSILVTVFLLLLSLPVLAGAITMLLTDRNFGTTFFAPDGGGDPVLFQHLFWFFGHPEVYILILPGFG MISQIVSTFSRKPVFGYLGMAYAMVAIGGIGFVVWAHHMYTVGMSSATQAYFVAATMVIAVPTGVKIFSW IATMWGGSIEFRAPMIWAVGFIFLFTVGGVTGVVLANAGVDRVLQETYYVVAHFHYVLSLGAVFAIFAGW YYWFPKMTGYMYNETLAKAHFWVTFIGVNLVFFPQHFLGLSGMPRRYVDYPDAFAGWNLVSSVGSYISGF GVLIFLYCVIDAFAKKVPAGDNPWGAGATTLEWTLPSPPPFHQFEVLPRVQ >gi|83594661|ref|YP_428413.1| Cytochrome c oxidase cbb3-type, subunit I [Rhodospirillum rubrum ATCC 11170] MTQATIARGARADTEPYVEGVIKKFVIAAVLWGVVGFIAGDVIAWQLAFPSLNMDLEWTSFGRLRPLHTS AVVFAFGGNVLLGTSLYVVQRTSRASLYGGAALGNIIFWGYQLFIVMAALGYVLGVTQGKEYAEPEWFVD LFLTVVWVLYLAAFLGTLLKRREPHIYVANWFFLAMIITIALLHLGNNMAIPVALMGGDSWVKSYGFYSG VQDAMTQWWYGHNAVGFFLTAGFLGIMYYFVPKQAQRPVYSYRLSIVHFWALIFLYIWAGPHHLHYTALP DWAQTVGMVFSVMLWMPSWGGMINGLMTLSGAWDKLRTDPVLRFLVVSVGFYGMSTFEGPMMSIKAVNSL SHYTDWTIGHVHSGALGWVAFVSFGALYYLVPALWKRRSLYSLKLVSLHFWIATLGIVLYITSMWVSGIM QGLMWRAYDELGFLQYSFIESVAAMHPYYIIRATGGVLFVIGSVVMVYNMYRTIKGDIREDAPQAAYLGS AAGVRR >gi|27377874|ref|NP_769403.1| cytochrome-c oxidase [Bradyrhizobium japonicum USDA 110] MSQPSISKSMTIGESGLAVVFAATAFLCVIAAAKALDAPFAFHAALSAAASVAAVFCIVNRYFERPAALP PAEINGRPNYNMGPIKFSSFMAMFWGIAGFLVGLIIASQLAWPALNFDLPWISFGRLRPLHTSAVIFAFG GNVLIATSFYVVQKSCRVRLAGDLAPWFVVVGYNFFILVAGTGYLLGVTQSKEYAEPEWYADLWLTIVWV VYLLVFLATIIKRKEPHIFVANWFYLAFIVTIAVLHLGNNPALPVSAFGSKSYVAWGGIQDAMFQWWYGH NAVGFFLTAGFLAIMYYFIPKRAERPIYSYRLSIIHFWALIFLYIWAGPHHLHYTALPDWTQTLGMTFSI MLWMPSWGGMINGLMTLSGAWDKLRTDPVLRMLVVSVAFYGMSTFEGPMMSIKVVNSLSHYTDWTIGHVH SGALGWVGFVSFGALYCLVPWAWNRKGLYSLKLVNWHFWVATLGIVLYISAMWVSGILQGLMWRAYTSLG FLEYSFIETVEAMHPFYIIRAAGGGLFLIGALIMAYNLWMTVRVGEAEVQMPVALQPAE >gi|146307622|ref|YP_001188087.1| cytochrome c oxidase, cbb3-type, subunit I [Pseudomonas mendocina ymp] MNTTTRSAYNYRVVRQFAIMTVVWGIVGMGLGVFIAAQLAWPDLNFNLPWTSFGRLRPLHTNAVIFAFGG CALFATSYYAVQRTSQTTLFAPKLAAFTFWGWQLVIVLAAISLPLGWTSSKEYAELEWPIDILITIVWVS YAIVFFGTVMQRKVSHIYVGNWFFGGFILTVAILHVVNNLEIPITLTKSYSLYAGATDAMIQWWYGHNAV GFFLTAGFLGMMYYFVPKQAGRPVYSYRLSIVHFWALIAVYIWAGPHHLHYTALPDWAQSLGMVMSLVLL APSWGGMINGMMTLSGAWHKLRTDPILRFLVVSLAFYGMSTFEGPMMAIKTVNALSHYTDWTIGHVHAGA LGWVAMVSIGSLYHLIPKVFGREQMHSIGLINSHFWLATIGTVLYIASMWVNGITQGLMWRAVNEDGTLT YSFVEALEASHPGFVVRVIGGAIFFAGMLLMAWNVWLTVRSAKSTEMEAAAQFSVEGAH Multiple sequence alignment We will use the sequences selected above as input in a multiple sequence alignment program (ClustalW). Download the program from http://inn-prot.weizmann.ac.il/software/ClustalX.html Information on the program can be found at: http://www.molbiol.ox.ac.uk/documentation/clustalx/clustalx.html Use the webinterface http://www.ebi.ac.uk/clustalw/ Align globally the sequences that you have selected. Test the influence of different parameters. Once you have obtained a reliable alignment: 1) Look at the alignment and the phylogenetic tree? What do you observe? 2) Compare the multiple alignment with the local pairwise alignments of two members of the family (e.g. dataset2). What is most informative the pairwise or the multiple alignment? 3) The sequences that you have added and that were not retrieved by the blast search? Do they belong in the multiple alignment? Why were they not detected by the blast search? 4) Change the parameters of the multiple alignment (different gap cost, lower), do you still find the right alignment? How sensitive is the finding the true alignment to the gap cost Results The system that will be studied is the terminal oxidase, the enzyme that catalyses the final reduction of O2 to H2O in the respiratory chain to generate energy. For more information see http://www.sanger.ac.uk/Software/Pfam/ The genes of which the sequences were aligned constitute Subunit 1 of the terminal oxidase complex. This subunit contains the catalytic site where O2 is reduced to H2O. It contains to this purpose a heme copper center. A conserved high spin and low spin heme are involved in ligating the Cu center. Conserved H residues have been shown to be involved in binding the heme. Three major families of terminal oxidases can be detected, 2 of which occur in prokaryotes only (cytcb3 type oxidases and quinol oxidases. The terminal oxidase all are part of the respiratory chain, they receive electrons from an electron donor and use these electrons to reduce the O2. Some terminal oxidases receive the elcttrons from quinols, other from cytochrome c. This explains the differences in sequence between the distinct classes of oxidases. Eukaryotes only have one type of oxidase: cytochrome c type oxidase: Prokaryotes often have branched repiratory chains with different type of terminal oxidases. Each of these oxidases has different properties (some are produced at very low O2 concentrations and have a high affinity for O2). This allows bacteria to live in very different environments while for eukaryotes a fixed O2 concentration is required. The cytochrome c type oxidases is the one that is also present in eukaryotes. For comparison the correct alignments with an indication on the structural important sites Multiple alignment: standard gap cost Ligands Cu center Ligands hemes