intron_retention_pro..
advertisement

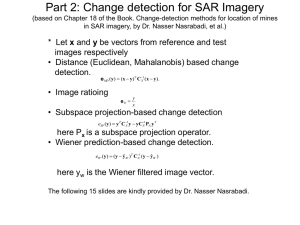
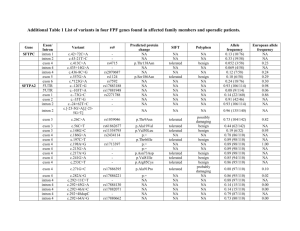
1) New sequence features. Since the gradation of GC content from flanking exons to intron may play an important role in separating retained introns from constitutive introns in plant species, I added new sequence features ratio of GC content from intron to its upstream and downstream exon which are all found to be discriminative. Frequency distribution and cumulative frequency distribution of these two features are shown in figure 1. 45000 120.00% 40000 100.00% 35000 Frequency 30000 IntronR frequency Cons. frequency IntronR cumulative% Cons. cumulative% 25000 20000 15000 80.00% 60.00% 40.00% 10000 20.00% 5000 0 0.00% 0.5 0.6 0.7 0.8 0.9 1 1.2 1.4 1.6 1.8 More Ratio of intron to upstream exon GC content Figure 1A. Ratio of intron to upstream exon GC content distribution. 45000 120.00% 40000 100.00% 35000 Frequency 30000 80.00% IntronR frequency Cons. frequency IntronR cumulative% Cons. Cumulative% 25000 20000 15000 60.00% 40.00% 10000 20.00% 5000 0 0.00% 0.5 0.6 0.7 0.8 0.9 1 1.2 1.4 1.6 1.8 More Ratio of intron to downstream exon GC content Figure 1B. Ratio of intron to downstream exon GC content distribution. 2) 11-fold cross validation in two approaches To test if we need to only train a single classifiers for whole plant species or to train species specific classifiers for each species individually, I applied RF to train a classifier to predict intron retention with 11 fold cross validation in two distinct approaches. Since the positive and negative datasets are unbalanced (19675 and 131353 respectively), I randomly chose the same number of negative dataset to match positive dataset while maintaining the proportion of data instances in each species. In the first approach, the training dataset was divided into 11 subsets by species. 10 subsets were used to train the classifier and the remaining one subset is used to test the classifier. The cross validation process then repeated 11 times with each of the subset used exactly once as the validation data. Results are shown in table 1. Fold 1 2 3 4 5 6 7 8 9 10 11 Species AT BD GM LJ MT OS PP PT SB SL VV Data instance 8462 142 3346 170 682 16676 4024 514 3342 272 1614 Total: 39244 AUC(20 trees) AUC(50 trees) 0.659 0.678 0.935 0.953 0.742 0.768 0.768 0.795 0.755 0.782 0.774 0.783 0.591 0.599 0.878 0.903 0.915 0.928 0.771 0.794 0.827 0.845 Weighted average: 0.7435 0.7574 Table 1. 11 fold species wide cross validation using Random Forest with 20 and 50 trees, each constructed while considering 9 random features. In the second approach, generic 11 fold cross validation was applied to train the classifier in which whole training set was divided into 11 subsets evenly instead of dividing by species. The average area under ROC curve (AUC) is 0.796 for 20 trees and 0.816 for 50 trees. Comparing to table 1, the weighted average AUC is notably higher in the second approach. In addition, the classifier performance is substantially lower than average for some species like Arabidopsis and Physcomitrella. Therefore, species specific classifiers are necessary for our study. 3) Species specific classifiers I applied RF to train our species specific classifiers base on 11 balanced datasets using 5 fold cross validation. We set 200 trees in RF, each constructed while considering 9 random features. Detailed accuracy for each classifier is shown in table 2. And ROC curves of 11 distinct classifiers are shown in figure 2. Species AT BD GM LJ Data instance 8462 142 3346 170 AUC (200 trees) 0.764 0.932 0.853 0.890 MT OS PP PT SB SL VV 682 16676 4024 514 3342 272 1614 Total: 39244 0.872 0.851 0.811 0.898 0.940 0.889 0.872 Weighted average: 0.838 Table 2. Performance evaluation of 11 species specific classifiers using RF. Figure 2. ROC curves of 11 distinct species specific classifiers using RF.