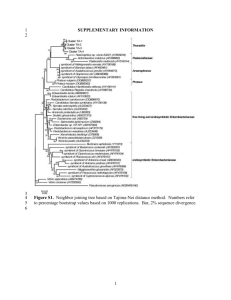
Supplementary Information (doc 42K)
advertisement

1 Supplementary Information 2 3 Table S1: Sampling details. Tube worm specimens were sampled during the two cruises I: 4 AT15-28 (“Fix08 I”, December 2007/January 2008) and II: AT15-38 (“Fix08 II”, October 5 2008) at different EPR vent sites. AD: Alvin Dive. The in situ chemistry measurements 6 (“Chemistry”) were performed during the cruise AT15-28 at 9°50’N, Tica vent. 7 8 Table S2: Single gene homogeneity. Selected genes with relevant metabolic functions and 9 the ITS and 16S rRNA sequences were compared a) between all three symbiont metagenomes 10 pyrosequenced in this study (R1: Riftia 1 symbiont, R2: Riftia 2 symbiont, T: Tevnia 11 symbiont) and b) between R1, R2, T and the Sanger-sequenced metagenome of 1: Cand. E. 12 persephone as published by Robidart et al. (2008), using the Geneious ProTM tool. 13 Homogeneity is given as percentage of average pairwise identities, i. e. the proportion of 14 homologous base pairs at the same column with regard to the total number of pairs in the 15 alignment. The homogeneity of all key metabolic gene sequences (excluding 16S rRNA and 16 ITS genes) from R1, R2 and T averages 99.9 %, resulting in an average heterogeneity of 0.1 17 %. The key gene alignment of R1, R2, T and the previously published Cand. E. persephone 18 metagenome resulted in an average of 99.6% homogeneity (0.4% heterogeneity). Five of the 19 genes were not identified in the Cand. E. persephone metagenome. For accession numbers see 20 Figures S2 and S3. 21 22 Table S3: Genes of particular interest. Selected representative genes encoding relevant 23 metabolic key enzymes, proteins involved in oxidative stress response and in cell surface- 24 associated processes, and proteins which are putatively related to symbiont-host interactions 25 are listed with their respective GenBank accession numbers for the metagenomes of the Riftia 2 1 1 symbiont, the Riftia 2 symbiont and the Tevnia symbiont. Asterisk: Protein is also involved 2 in organic carbon metabolism. 1) Two or more partial coding sequences for one gene; 2) two or 3 more individual copies in separate locations of the metagenomes. n. a.: “not annotated”, 4 nucleotide sequence was manually identified in the metagenome but was not detected during 5 the initial automatic annotation. (Note: This list is not exhaustive but presents exemplary 6 features of the three metagenomes.) 7 8 Table S4: Comparison of intracellular soluble proteins. Protein names with corresponding 9 function, Enzyme Commission (E.C.) number, isoelectric point (pI), molecular weight (MW) 10 and GenBank accession number are listed for all identified proteins with at least 1.5-fold 11 change of spot volume (ratio) when comparing the Riftia symbiont and the Tevnia symbiont 12 protein gels (Figure 2). Spot volume values are expressed in percentages (%Vol) of the total 13 proteome on the respective symbiont reference map (pI range 4 – 7). Only proteins exhibiting 14 relative volumes (%Vol) of at least 0.1 were included. Negative ratio values (light gray cells) 15 correspond to comparatively higher spot volumes on the Tevnia symbiont master gel, whereas 16 positive ratios (dark gray cells) indicate larger spot volumes for the Riftia symbiont proteins. 17 Proteins were considered as unambiguously identified, if their MS identification was based on 18 at least two individual peptides, a minimum score of 75 and a sequence coverage of at least 19 30%. Asterisk: Protein is also involved in organic carbon metabolism. 20 21 Figure S1: Tube worm clump at the sampling site. Tevnia jerichonana and Riftia pachyptila 22 specimens at Tica vent, 9°50’N. The picture was taken on January 11th 2008 directly before 23 collection of the tube worms. The long red plumes of the larger Riftia specimens in the center 24 of the clump are more distant from the diffuse flow source at the seafloor than the plumes of 25 the smaller Tevnia specimens. 2 1 2 Figure S2: Single gene comparison. 24 key genes encoding major metabolic enzymes 3 involved in sulfide oxidation, carbon fixation, nitrogen metabolism and oxidative stress 4 response from the Riftia 1 symbiont, the Riftia 2 symbiont, and the Tevnia symbiont 5 metagenome were subjected to a mutual alignment and compared to the respective Riftia 6 symbiont sequences published previously (Robidart et al., 2008) using the Geneious ProTM 7 tool. The green bar indicates the nucleotide consensus sequence (shown on top), i. e. the 8 homogeneity of the four symbiont metagenomes. It is interrupted in differing (non- 9 homologous) DNA regions. Protein coding sequences (CDS) of the respective symbionts are 10 indicated by a yellow bar and specified by their GenBank accession numbers on the left. 11 Translations (above the DNA sequences) indicate whether nucleotide deviations impact the 12 amino acid sequences. 13 14 Figure S3: ITS and 16S rRNA alignment. ITS and 16S rRNA sequences of the three 15 metagenomes sequenced in this study (Riftia 1 symbiont, Riftia 2 symbiont, Tevnia symbiont) 16 were compared to each other and to the sequences previously published by Robidart et al. 17 (2008) using Geneious ProTM. Additionally, ITS sequences of two individual Riftia symbiont 18 phylotypes described by Harmer et al. (2008) and of seven Riftia and Tevnia symbiont 19 phylotypes as published by Di Meo et al. (2000) were included in the alignment. The 16S 20 rRNA alignment also includes two 16S rRNA sequences published by Di Meo et al. (2000). 21 GenBank accession numbers of publically accessible sequences are indicated on the left. The 22 pink bar characterizes tRNA sequences located within the ITS sequence, whereas the rRNA 23 sequence is shown as a red bar. The consensus sequence (homogeneity of the compared 24 genes) is highlighted in green. 3