Supplementary Figure and Table Legends (doc 41K)
advertisement

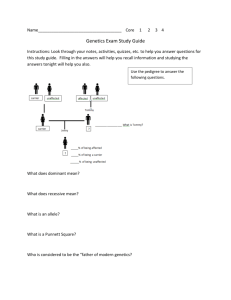
1 Supplementary Figure and Table Legends 2 3 Suppl. Figure 1 4 Workflow for analysis of exome sequence data. Variant prioritisation criteria: (i) 5 variants were non-synonymous (includes nonsense, frameshift and missense) or splice 6 site variants, (ii) variants did not occur with a frequency of ≥ to 1% in 1000genomes, 7 ESP5400 or dbSNP (unless they were a previously defined pathogenic blindness variant 8 in dbSNP), (iii) variants were covered by at least 10 reads with at least 20% variation 9 reads or at least 5 reads with at least 40% variation reads, and (iv) variants were 10 considered “probably pathogenic” on the basis of the type of mutation and our in silico 11 analysis criteria. 12 13 14 Suppl. Figure 2 15 Pedigrees and Sanger sequence analysis in families with pathogenic variants and 16 segregation indicating autosomal dominant or recessive inheritance patterns. Variants 17 are indicated by a blue vertical line. 18 Family 1: De novo heterozygous mutation in GJA8 in II.1 (Patient 1), not present in her 19 parents and passed onto the offspring in an autosomal dominant manner. 20 Family 4: Heterozygous mutation in CRYGC in I.2 (Patient 4), passed onto the children 21 in an autosomal dominant manner. 22 Family 5: Compound heterozygous mutations in CYP1B1 in II.1 (Patient 5) that have 23 been inherited in an autosomal recessive manner from each of her unaffected parents 24 who are heterozygous for each of the mutations. 1 25 Family 6: De novo heterozygous mutation in PAX6 in II.1 (Patient 6), and not present in 26 either of her parents. 27 Sequences are shown in forward direction, apart from CRYGC sequence in Family 4 28 which is in reverse. + symbol indicates a normal, non-variant allele. Reference 29 sequences 30 NM_000104.3; PAX6, NM_000280.4. used: GJA8, NM_005267.4; CRYGC, NM_020989.3; CYP1B1, 31 32 Suppl. Figure 3 33 Pedigrees and Sanger sequence analysis in families with variants with incomplete 34 penetrance or inheritance pattern not consistent with the disease phenotype. 35 Family 3: Heterozygous variant in CRYBA1 in II.1 (Patient 3), also present in her 36 unaffected mother. 37 Family 9: Compound heterozygous variants in CYP1B1 in II.1 (Patient 9), that have 38 been inherited in an autosomal recessive manner from each of her unaffected parents 39 who are heterozygous. Her affected brother is heterozygous for one of the CYP1B1 40 variants. Heterozygous variant in BFSP1 in II.1 (Patient 9), that is also present in her 41 affected brother and her unaffected mother. 42 Family 10: Heterozygous variant in ABCB6 in II.1 (Patient 10), which is also present in 43 his unaffected father. Heterozygous variant in SLC16A12 in II.1 (Patient 10), which is 44 also present in his unaffected mother. 45 Family 11: Heterozygous variant in GDF3 in II.1 (Patient 11), which is also present in 46 his unaffected father. Compound heterozygous variants in CYP1B1 in II.1 (Patient 11), 47 that have been inherited in an autosomal recessive manner from each of his unaffected 2 48 parents who are heterozygous. Heterozygous variant in BFSP1 in II.1 (Patient 11), 49 which is present in his unaffected mother and his unaffected father. 50 Sequences are shown in forward direction, apart from CRYBA1 sequence in Family 3, 51 SLC16A12 sequence in Family 10, and GDF3 and BFSP1 sequences in Family 11 52 which are in reverse. + symbol indicates a normal non-variant allele. 53 Reference sequences used: CYP1B1, NM_000104.3; CRYBA1, NM_005208.4; BFSP1, 54 NM_001161705.1; ABCB6, NM_005689.2; SLC16A12, NM_213606.3; and GDF3, 55 NM_020634.1 56 57 Suppl. Table 1 58 56 developmental eye disease genes examined in this study. 59 60 Suppl. Table 2 61 Clinical ocular phenotypes in patients undergoing exome sequencing and family 62 members. 63 64 Suppl. Table 3 65 Exome sequencing analysis and variant prioritisation for developmental eye disease 66 gene identification. 3