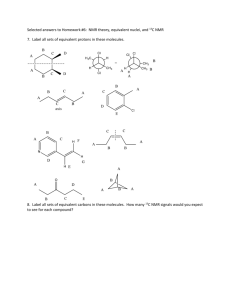
Supplementary Information (doc 134K)
advertisement
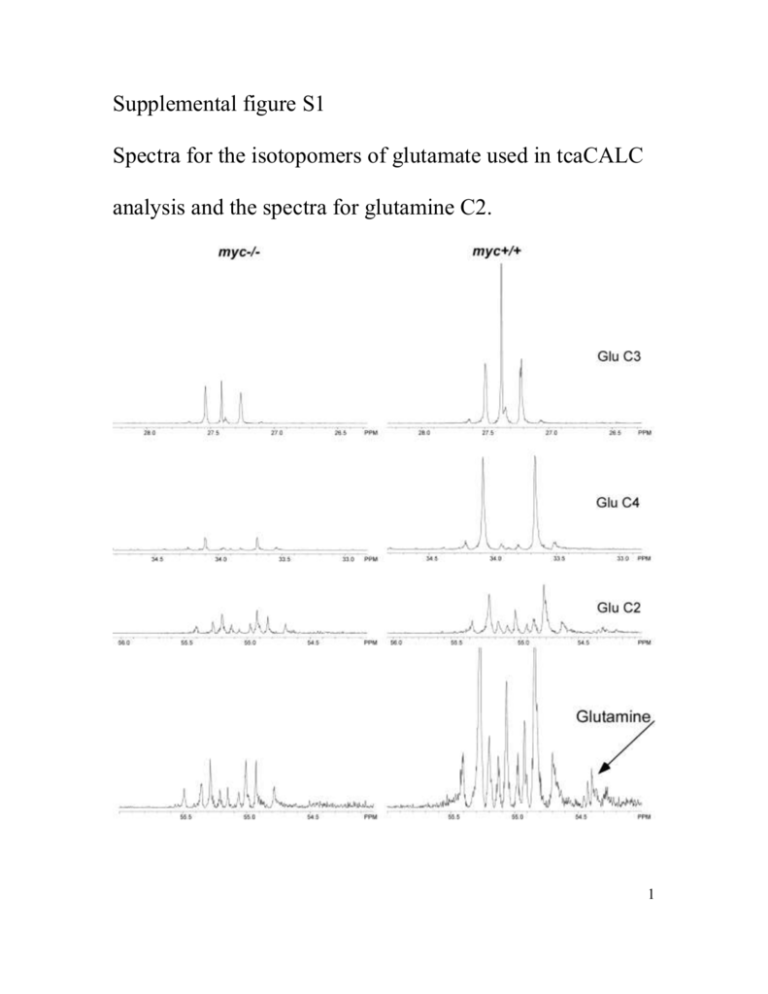
Supplemental figure S1 Spectra for the isotopomers of glutamate used in tcaCALC analysis and the spectra for glutamine C2. 1 Supplemental methods Cell lines and reagents The cell lines used include the TGR-1 subclone of the Rat-1 fibroblast cell line (myc+/+) and myc-/- and myc-/-MycER derivatives of TGR-1 as described (O'Connell et al, 2003). [U-13C] labeled glucose was purchased from Cambridge Isotopes (Andover, MA), methotrexate and 6-diazo-5-oxo-L-norleucine from Sigma. Immunoblot analysis Extracts were prepared as described (Slawson et al., 2005) and after SDS-PAGE, blotted and probed with the CTD 110.6 antibody (Covance, Berkeley), Anti-O-GlcNAc transferase (DM-17, Sigma) and tubulin (Oncogene, Biosciences ) . Cell culture Cells were plated in 10% serum and grown for 48 hours until confluent, media was replaced with 0.25% bovine calf serum (BCS;InVitrogen) and cells grown a further 48 hours. Cells were then split and re-plated in 10% BCS for 12 hours. After washing with PBS, cells were incubated in HEPES buffered saline (HBSS;114 mM NaCl/4.7 mmol KCl/1.2 mM KH2PO4/1.16 mM MgSO4/20 mM Hepes/2.5 mM CaCl2/25.5 mM NaHCO3, pH 7.2) in the presence of 10% dialyzed fetal calf serum (Hyclone) and 20 mM [U-13C] glucose (Cambridge Isotopes) for 4 hours. Cells were washed, harvested by trypsinization, washed, pelleted by centrifugation, and flash frozen in liquid nitrogen. Cell extracts for NMR were prepared using chloroform methanol as described (McGrath 2 et al, 2006). Extracts from equivalent numbers of cells were lyophilized and deutriated water (Sigma) added prior to analysis by NMR. Cell growth assays were performed by HO33342 staining as described previously (Morrish et al., 2008). NMR 13 C NMR spectra were acquired at 500 MHz and 20 0C on a Varian Unity-500 NMR spectrometer equipped with a 5 mm broadband “switchable” probe (Varian, Palo Alto, CA). The acquisition parameters were as follows: radio frequency pulse angle of 450, spectral width of 27.6 kHz, acquisition time of 1.185 s and 3 s repetition period between consecutive scans. Proton decoupling was achieved using a Waltz decoupling pulse sequence. The signal-to-noise ratio was enhanced by a 5 Hz exponential multiplication function. TMPSA (1 mM) was used as an internal reference. Appropriate correction factors for nuclear Overhauser enhancement (NOE) and signal saturation were determined. Spectra were processed with one left shift, 2 levels of zero fill prior to Fourier transformation. 13C NMR multiplets were deconvoluted using NUTS (Acorn NMR Freemont, CA). Concentrations of 13C labeled metabolites were calculated by comparison to the relative integral of the TMPSA standard (1mM), following subtraction of natural abundance integral values. To confirm spectral assignments heteronuclear single quantum correlation (HSQC Varian pulse program , with TD (F2)= 1024 (512 complex pairs), TD(F1)= 512 (256 complex pairs), SW (1H)= 11 ppm, SW (13C)=180 ppm, GARP decoupling NS=128, AQ=0.78 s, D1=1.2 s, States-TPPI (phase = 1,2) hypercomplex phase cycling, and heteronuclear multiple bond correlation (HMBC Varian pulse program optimized for long range couplings, TD(F2)= 2048 (1024 complex pairs), 3 TD(F1)=800 (400 complex pairs), NS=128, D1= 1.2s, SW (1H)=11ppm, SW(13C)=210ppm, AQ=0.155 s, GARP decoupling experiments were performed to correlated 1H and 13C resonances. These spectra were acquired on a 600 MHz spectrometer interfaced with a 14.1 T superconducting magnet using a Varian Cold ProbeTM. LC/MS/MS Glutamic acid was analyzed using mass spectrometry in the form of butyl ester (Chace et al., 1993). For improved signal to noise ratio and to remove tryptophane interference a short reverse phase LCMS method was used. Agilent 1100 HPLC with Zorbax SB-1 C18 column (2.1x100mm, 80A pores and 3.6um particles) was coupled to EsquireLC ion trap mass spectrometer operated in positive ionization mode. Water with 5% acetonitrile, 1% acetic acid and acetonitrile with 1% acetic acid were used as solvents A and B respectively. Gradient from 10% B to 64% B in 9 minutes was followed by 5min at 100%B and column was equilibrated during 12 minutes at a flow rate of 200 uL/min. Glutamic acid retention time was 8.5 minutes. Quantitative isotopic incorporation was analyzed by deconvolution of the experimental isotope pattern using linear combination of contributions of unlabeled and all theoretically possible 13 C labeled glutamate molecules. The percent value of individual contributions was iteratively optimized to match the experimental isotope pattern using Microsoft Excel. Metabolic analysis 4 Metabolic substrate provision to the TCA from different pathways was estimated from the relative areas of the multiplets of glutamate C2, C3 and C4 in the 13C NMR spectra using tcaCALC (Malloy et al., 1990). The 13C NMR data was fitted to a standard model of catabolic mammalian cell metabolism. The model parameters that were considered were: (1) the fraction of acetyl-CoA derived from the oxidation of [U-13C] pyruvate via pyruvate dehydrogenase; (2) the fractional contribution of [U-13C] pyruvate to oxaloacetate via pyruvate carboxylase; (3) the fraction of acetyl-CoA derived from unlabeled acyl sources; (4) relative pyruvate carboxylase and pyruvate dehydrogenase flux. All flux parameters are relative to that of citrate synthase, arbitrarily set at 1.0. The standard error associated with each parameter estimate was calculated by Monte Carlo simulation. Fittings were performed systematically until the experimental and calculated 13 C multiplet data showed the best fit. Statistical analysis Data are expressed as means +/- S.D. values for the indicated number of experiments. Statistical comparisons were made using student T test and a p-value <0.05 was considered statistically significant. References Chace, D.H., Millington, D.S., Terada, N., Kahler, S.G., Roe, C.R. and Hofman L.F. (1993) Rapid diagnosis of phenylketonuria by quantitative analysis for phenylalanine and tyrosine in neonatal blood spots by tandem mass spectrometry. Clin Chem. 39, 66-71. 5 McGrath, B.M., Greenshaw, A.J., McKay, R., Slupsky, C.M., and Silverstone, P.H. (2006). Lithium alters regional rat brain myo-inositol at 2 and 4 weeks: an ex-vivo magnetic resonance spectroscopy study at 18.8 T. Neuroreport 17, 1323-1326. Malloy, C.R., Sherry, A.D., and Jeffrey, F.M. (1990). Analysis of tricarboxylic acid cycle of the heart using 13C isotope isomers. Am J Physiol 259: H987-95. Morrish, F.M., Neretti, N. Sedivy, J.M. and Hockenbery, D. M. (2008) The oncogene cMyc coordinates regulation of metabolic networks to enable rapid cell cycle entry. Cell Cycle 7: 1054-1066. O'Connell, B.C., Cheung, A.F., Simkevich, C.P., Tam, W., Ren, X., Mateyak, M.K., and Sedivy, J.M. (2003). A large scale genetic analysis of c-Myc-regulated gene expression patterns. J Biol Chem 278, 12563-73. 6