Microsoft word version
advertisement

Tutorial III
LOD Scores, mQTLs and SNP Marker Analysis
Steve Horvath, Anatole Ghazalpour, Sud Doss, Bin Zhang
Correspondence: shorvath@mednet.ucla.edu,
http://www.ph.ucla.edu/biostat/people/horvath.htm
Abstract
This tutorial is the direct sequel of tutorial II
The tutorials and data files can be found at the following webpage:
http://www.genetics.ucla.edu/labs/horvath/CoexpressionNetwork/MouseWeight/
More material on weighted network analysis can be found here
http://www.genetics.ucla.edu/labs/horvath/CoexpressionNetwork/
The data and biological implications are described in
Anatole Ghazalpour, Sudheer Doss, Bin Zang, Susanna Wang,Eric E. Schadt,
Thomas A. Drake, Aldons J. Lusis, Steve Horvath (2006) Integrating Genetics and
Network Analysis to Characterize Genes Related to Mouse Weight. PloS Genetics.
We provide R code that shows
a) how to relate clinical traits and expression profiles to genetic markers
Quantitative trait locus, LODscore, QTL, linkage analysis, allelic association
analysis, mQTL
b) how to produce Figure 3 in our article (LOD score curve and mQTL graph)
c) how to regress a phenotype on multiple markers with the fitqtl package
(Broman et al 2003).
The purpose of this tutorial is to demonstate how the body weight QTL, expression QTLs
and Module QTLs (mQTLs) were calculated.
We make use of the R/qtl package (Broman et al 2003) which is explained in Dr
Broman’s tutorial at the following webpage.
http://www.biostat.jhsph.edu/~kbroman/qtl/rqtltour.pdf
To avoid duplication of effort, this tutorial is rather terse with regard to QTL analysis.
Weight QTL analysis
For bodyweight QTL analysis, in summary, we first read the genotype and phenotype
data into the R software using read.cross function (make sure the R working directory is
set to the correct directory):
setwd("C:/Documents and Settings/shorvath/My
Documents/ADAG/AnatoleGhazalpour/MOUSEWEIGHT/TUTORIALS/ThirdTutorial")
library(qtl)
# now we read in the genetic marker data from the F2 mouse cross (female BXHxC3H
F2crossdata <- read.cross("csvr","", "F2mouseBXHxC3HGenotypes.csv")
1
# The following data frame contains the LOD scores for each gene (expression) across
# the SNP markers.
ExpressionLodScore=read.csv("ExpressionLODscoreFemaleLiver.csv",header=T)
SNPtable=read.csv("SNPMarkerLocusTranslationTable.csv")
colorh1=ExpressionLodScore$ModuleFemaleLiver
#We defined a module quantitative trait locus (mQTL) as the locus with a significant
#enrichment for eQTL of the genes within a predetermined gene module.
# Here we specify the module of interest
whichmodule="blue"
LODscoreThreshold=2
NoSignifModuleGenes=apply(ExpressionLodScore[colorh1==whichmodule, -c(1:3)]>
LODscoreThreshold,2,sum)
NoSignifGenes=apply(ExpressionLodScore[, -c(1:3)]> LODscoreThreshold,2,sum)
par(mfrow=c(2,1) )
plot(NoSignifGenes,type="h", col="black", xlab="Marker Number",main=paste("No. of
network genes with epression LOD score >", LODscoreThreshold) )
plot(NoSignifModuleGenes,type="h", col=whichmodule, xlab="Marker Number", main=
paste("No of", whichmodule, " module genes with expression LOD score >",
LODscoreThreshold))
# This data frame contains the no. of significant genes for each SNP (rows)
NoSignifGenesData=data.frame(UCSC.Name=names(NoSignifModuleGenes),
NoSignifModuleGenes= as.vector(NoSignifModuleGenes), NoSignifGenes=
as.vector(NoSignifGenes) )
# This outputs the SNPs with more for which more than 100 module genes
# have a LOD score > LODscore threshold
NoSignifGenesData[NoSignifGenesData$ NoSignifModuleGenes>100,]
2
UCSC.Name NoSignifModuleGenes NoSignifGenes
112
p45066
117
512
113
p46356
117
512
114
p45960
117
512
3
#To calculate the weight QTL we first run a genome scan using the scanone function:
F2crossdata <- calc.genoprob(F2crossdata, error.prob=0.01)
F2crossdata <- calc.genoprob(F2crossdata, error.prob=0.01)
out.hk <- scanone(F2crossdata, method="hk")
#To view the highest LOD scores for each chromosome type:
summary(out.hk)
p45504
p45236
p44989
p44990
p45558
p44960
p45323
p45103
p45722
p45653
p45738
p45039
p46019
p45050
p44593
p45947
p45618
p44804
p45500
chr
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
pos
142.7
144.3
160.0
155.2
118.5
125.6
136.7
13.7
36.3
40.6
9.4
79.6
71.0
116.8
78.2
3.6
26.4
73.8
50.4
lod
3.03
0.63
1.85
1.03
3.39
0.36
1.07
0.70
1.55
0.96
1.02
0.87
1.16
0.73
3.58
1.28
2.13
1.02
4.07
AA
39
39
36
36
35
38
38
38
40
38
37
40
36
40
41
40
35
40
40
AB
39
38
38
39
39
39
39
39
37
37
38
38
39
37
38
37
39
37
39
BB sigma
34
5.8
37
6.1
40
5.9
38
6.0
41
5.8
37
6.1
36
6.0
37
6.0
38
6.0
40
6.0
40
6.0
37
6.0
38
6.0
38
6.0
36
5.7
38
6.0
39
5.9
38
6.0
35
5.7
4
#To view the LOD score curve of body weight
par(mfrow=c(1,1))
plot(out.hk,gap=0,col="black")
5
# Now we merge the 2 data sets
datmerge=merge(NoSignifGenesData, data.frame(UCSC.Name=
dimnames(out.hk)[[1]], out.hk), by.x="UCSC.Name", by.y="UCSC.Name",sort=F)
dim(NoSignifGenesData)
dim(out.hk)
dim(datmerge)
names(SNPtable)
datmerge=merge(SNPtable, datmerge, by.x="UCSC.Name",
by.y="UCSC.Name",sort=F)
names(datmerge)
dim(datmerge)
attach(datmerge)
6
Since we were interested in studying the genetics of modules as opposed to the genetics
of the entire network, when searching for mQTLs, we focused on module-specific eQTL
hot spots. Toward this end, we used the Fisher exact test to determine whether the
proportion of module genes that map to the eQTL hotspot was significantly higher than
that of the 8,000 network genes. While we used the 8000 most varying genes in our
paper, we only have 3421 in this tutorial. The following code shows how to pick mQTLs
based on the 3421 genes. Naturally, the results will be slightly different from the analysis
reported in Table 1 of the paper since it was based on the 8000 genes.
# For the i-th SNP,
# the following table reports the number of significant genes (first column) versus
# non-significant genes (second column) for the module genes (first row)
# and the non-module genes (second row).
fisherpvalue=rep(NA, length(NoSignifGenes) )
for (i in c(1: length(NoSignifGenes) ) ) {
tab1=matrix(NA, nrow=2,ncol=2)
tab1[1,1]= NoSignifModuleGenes[i]
tab1[1,2]=sum(colorh1==whichmodule)- tab1[1,1]
tab1[2,1]= NoSignifGenes[i]- NoSignifModuleGenes[i]
tab1[2,2]= length(colorh1)- sum(colorh1==whichmodule)-tab1[2,1]
fisherpvalue[i]=fisher.test(tab1)$p.value
}
# This is the proportion of the number of significant module genes by no. of signif. genes.
enrich1=NoSignifModuleGenes/ NoSignifGenes
#To select the mQTLs based on the 3421 genes (not the 8000 genes mentioned in the
paper), we use the following 3 criteria
# this ensures that the eQTL hotspot (mQTL) is module specific (2 sided test)
1) fisherpvalue<0.0005
# This ensures that at least 20% of the significant genes are in the module at the mQTL
2) enrich1>.2
# this requires that at least 30 module genes map to the mQTL (LOD score threshold 2)
3) NoSignifModuleGenes>=30
selectmQTL=fisherpvalue<0.0005 & enrich1>.2 & NoSignifModuleGenes>=30
7
datout=data.frame(chromo=datmerge[,3],
datmerge[,c(1:2,7,8)],FisherP=signif(fisherpvalue,2), Enrich=signif(enrich1,2)
)[selectmQTL,]
# As can be seen from following table, multiple SNPs on the same chromosome satisfy
#the mQTL selection criteria. This is to be expected since SNPs that are close tend to be
#in linkage disequilibrium. In our paper, we chose a single representative (highlighted in
yellow) of every region.
> datout
chromo UCSC.Name Celera.Name NoSignifModuleGenes NoSignifGenes FisherP Enrich
49
1
p44629
rs3694065
34
119 2.5e-04
0.29
50
1
p44669
rs3717264
37
112 3.5e-06
0.33
51
1
p44901
rs3664662
37
111 3.1e-06
0.33
52
1
p45733
rs3658965
37
112 3.5e-06
0.33
53
1
p46102
rs3715814
37
121 3.0e-05
0.31
54
1
p45343
rs3725808
35
106 7.3e-06
0.33
55
1
p45507
rs3719973
34
98 2.7e-06
0.35
56
1
p45920
rs3659590
39
110 1.7e-07
0.35
57
1
p45182
rs3717377
39
110 1.7e-07
0.35
58
1
p45788
rs3677375
38
108 4.7e-07
0.35
59
1
p45339
rs3678662
38
108 4.7e-07
0.35
112
2
p45066
rs3655184
117
512 2.5e-06
0.23
113
2
p46356
rs3662347
117
512 2.5e-06
0.23
114
2
p45960
rs3707138
117
512 2.5e-06
0.23
188
3
p45967
rs4223971
30
64 2.6e-09
0.47
193
3
p45694
rs3715204
32
79 6.1e-08
0.41
194
3
p45241
rs3714575
54
110 5.2e-17
0.49
195
3
p46112
rs3717453
54
110 5.2e-17
0.49
196
3
p46111
rs3680433
54
110 5.2e-17
0.49
197
3
p46033
rs3671511
54
110 5.2e-17
0.49
198
3
p45546
rs3714671
55
115 1.2e-16
0.48
199
3
p44944
rs3705680
55
115 1.2e-16
0.48
200
3
p45317
rs3706170
33
73 1.3e-09
0.45
201
3
p45911
rs3707356
33
73 1.3e-09
0.45
202
3
p45620
rs3704498
33
73 1.3e-09
0.45
230
3
p46075
rs3698974
35
115 4.0e-05
0.30
231
3
p45544
rs3679962
35
115 4.0e-05
0.30
330
5
p46116
rs3688942
37
117 1.1e-05
0.32
331
5
p46327
rs3713492
37
117 1.1e-05
0.32
332
5
p45414
rs3713195
31
105 2.9e-04
0.30
333
5
p46249
rs3707624
34
115 1.1e-04
0.30
334
5
p45879
rs3679606
34
115 1.1e-04
0.30
335
5
p45249
rs3711950
36
118 4.3e-05
0.31
336
5
p44714
rs3656989
33
106 5.3e-05
0.31
337
5
p46273
rs3671517
35
122 1.8e-04
0.29
338
5
p44953
rs3684754
35
122 1.8e-04
0.29
339
5
p45322
rs3718145
37
126 7.7e-05
0.29
340
5
p46130
rs4138743
37
123 3.7e-05
0.30
341
5
p45840
rs3672514
37
126 7.7e-05
0.29
347
5
p44956
rs3721607
44
162 1.3e-04
0.27
348
5
p45839
rs3667334
44
162 1.3e-04
0.27
349
5
p44954
rs3688276
44
162 1.3e-04
0.27
350
5
p46184
rs3667067
44
162 1.3e-04
0.27
351
5
p45247
rs3686635
44
162 1.3e-04
0.27
352
5
p45750
rs3686672
44
162 1.3e-04
0.27
353
5
p45561
rs3716445
44
162 1.3e-04
0.27
612
10
p46016
rs3659700
34
76 1.0e-09
0.45
613
10
p46367
rs3660611
34
76 1.0e-09
0.45
614
10
p46411
rs3708275
39
99 5.3e-09
0.39
615
10
p46210
rs3673471
38
73 2.6e-13
0.52
616
10
p45930
rs3708450
40
80 3.4e-13
0.50
617
10
p46344
rs3693867
46
108 8.1e-12
0.43
618
10
p45651
rs3680457
42
92 4.3e-12
0.46
619
10
p44569
rs3677768
42
91 2.7e-12
0.46
620
10
p45928
rs3688363
54
91 4.1e-22
0.59
621
10
p45139
rs3679120
39
84 1.4e-11
0.46
8
622
623
625
626
627
628
629
630
631
632
633
637
638
705
706
707
708
709
710
711
712
713
714
715
722
723
724
725
726
727
728
729
730
731
732
733
756
766
767
768
769
824
825
826
827
828
829
830
831
1047
1048
1049
1050
1051
1052
1053
1054
1055
1056
1057
1058
10
10
10
10
10
10
10
10
10
10
10
10
10
12
12
12
12
12
12
12
12
12
12
12
12
12
12
12
12
12
12
12
12
12
12
12
12
13
13
13
13
14
14
14
14
14
14
14
14
19
19
19
19
19
19
19
19
19
19
19
19
p45511
p45653
p45025
p45347
p44731
p44673
p45023
p45797
p44526
p44560
p46293
p45026
p45863
p44633
p45364
p45517
p45659
p45588
p45144
p45363
p45362
p45464
p45143
p44635
p46266
p46225
p45126
p45194
p46054
p46227
p46219
p45358
p45937
p45356
p45802
p45196
p45801
p45369
p45438
p45664
p45523
p46296
p45807
p45207
p45439
p44590
p44591
p46384
p44919
p46074
p45826
p46355
p46124
p46123
p45229
p45500
p44890
p44892
p45916
p45915
p44811
rs3672342
rs3719045
rs3696307
rs3682060
rs3676909
rs3723140
rs3661840
rs3690226
rs3679452
rs3667599
rs3721306
rs3704401
rs3694833
rs3653990
rs4138084
rs3678128
rs3709002
rs3707458
rs3708624
rs3717933
rs3693390
rs3658504
rs3694821
rs3709287
rs3689251
rs3669704
rs3695382
rs3658204
rs3661834
rs3716646
rs3683481
rs3671194
rs3686437
rs3655309
rs3663333
rs3663603
rs3663172
rs3711424
rs4139503
rs3720707
rs3707097
rs3681670
rs3153444
rs3707560
rs3688833
rs3691821
rs3710549
rs3666026
rs3721918
rs3653886
rs3697994
rs3697139
rs3677115
rs3656289
rs3658160
mCV22979960
mCV22979964
mCV23068522
mCV23069037
mCV22608007
rs3722335
56
47
49
62
62
35
31
34
31
31
31
34
32
52
44
44
33
33
33
33
33
37
37
35
31
32
32
32
32
51
51
47
47
31
31
34
40
39
39
39
49
44
55
64
64
75
63
63
58
51
50
50
50
54
71
49
49
34
34
34
34
91
81
135
145
145
79
77
92
77
77
77
98
102
109
81
81
68
68
68
68
65
83
83
89
60
68
68
68
68
101
101
98
98
64
64
60
117
108
108
108
126
71
90
100
100
132
116
109
99
75
67
67
67
80
105
78
78
57
57
57
56
5.0e-24
8.8e-19
1.6e-09
1.0e-15
1.0e-15
7.9e-10
1.2e-07
4.6e-07
1.2e-07
1.2e-07
1.2e-07
2.7e-06
4.4e-05
9.7e-16
3.7e-16
3.7e-16
1.2e-10
1.2e-10
1.2e-10
1.2e-10
2.6e-11
2.0e-10
2.0e-10
3.5e-08
5.9e-11
6.5e-10
6.5e-10
6.5e-10
6.5e-10
8.8e-17
8.8e-17
2.0e-14
2.0e-14
4.9e-10
4.9e-10
1.9e-13
3.8e-07
9.9e-08
9.9e-08
9.9e-08
8.8e-11
3.2e-19
2.1e-23
7.8e-29
7.8e-29
8.0e-29
8.6e-23
8.2e-25
2.7e-23
6.1e-25
1.6e-27
1.6e-27
1.6e-27
4.0e-26
2.5e-34
1.1e-21
1.1e-21
2.3e-14
2.3e-14
2.3e-14
1.1e-14
0.62
0.58
0.36
0.43
0.43
0.44
0.40
0.37
0.40
0.40
0.40
0.35
0.31
0.48
0.54
0.54
0.49
0.49
0.49
0.49
0.51
0.45
0.45
0.39
0.52
0.47
0.47
0.47
0.47
0.50
0.50
0.48
0.48
0.48
0.48
0.57
0.34
0.36
0.36
0.36
0.39
0.62
0.61
0.64
0.64
0.57
0.54
0.58
0.59
0.68
0.75
0.75
0.75
0.68
0.68
0.63
0.63
0.60
0.60
0.60
0.61
Comment: Note that this analysis involving the 3421 genes suggests to define mQTLs for
chromosomes 1 and 13 as well. This could lead to an integrated models (see tutorial IV)
that provide an even better fit than the one reported in our paper. But here we will
proceed with the mQTLs defined in our paper, i.e we proceed with the following SNPs
9
mQTLnames=c("rs3662347","rs3714671","rs3721607","rs3676909", "rs3704401",
"rs3658504","rs3683481","rs3691821","rs3658160")
# this vector indicates the locations of the mQTLS used in our paper
LocationmQTL=is.element(Celera.Name, mQTLnames)
# This vector assigns color red to mQTLs and black for other SNPs
ColormQTL=ifelse(LocationmQTL,"red","black")
10
# Here we compare the number of module eQTLs to the the total number of eQTLs
par(mfrow=c(1,1))
jitterNoSignifGenes=jitter(NoSignifGenes,amount=8)
plot(NoSignifModuleGenes, jitterNoSignifGenes,xlab="No. Signif *Module* Genes at a
SNP", ylab="No. Signif Genes at a SNP", type="n",cex=.75)
text(NoSignifModuleGenes, jitterNoSignifGenes, labels=Celera.Chromosome
,col=ColormQTL,cex=.75)
lm1=lm(NoSignifGenes~ poly( NoSignifModuleGenes,2))
x= seq(from=min(NoSignifModuleGenes),to= max(NoSignifModuleGenes) ,
length.out=1000)
modelprediction = predict(lm1, data.frame(NoSignifModuleGenes=x))
lines(x, modelprediction)
# The SNPs in the plot above are labelled by chromosome location. mQTLs are colored
in red.
11
# Here we compare the number of module eQTLs to the the total number of eQTLs
par(mfrow=c(1,1))
plot(NoSignifModuleGenes[LocationmQTL],
NoSignifGenes[LocationmQTL],xlab="No. Signif *Module* Genes at a SNP",
ylab="No. Signif Genes at a SNP", main="mQTL only", type="n")
text(NoSignifModuleGenes[LocationmQTL], NoSignifGenes[LocationmQTL],
col="red", labels= Celera.Chromosome[LocationmQTL])
lines(x, modelprediction)
mQTL only
400
300
200
5
10
100
No. Signif Genes at a SNP
500
2
12
10
3
14
19
12
40
60
80
100
120
No. Signif *Module* Genes at a SNP
12
# As a pre-processing step for producing Figure 3, we create a position score (pos1) for
#the x-axis.
# the following code assumes that the chromosomes are sorted in order 1-19
# and that pos1 reports the marker distance on each chromosome
pos1=pos
chr1=as.numeric( chr)
tickmarkposition1=rep(NA,length(unique(chr1)))
chromonumber=1
tickmarkposition1[1]=pos1[chromonumber]
for (i in c(2:(length(chr1)-1) ) ) {
if (chr1[i] !=chr1[i-1] ) {
chromonumber=chromonumber+1
gapbetweenchromosomes=30
pos1[i:length(chr1)]= pos1[i:length(chr1)]+pos1[i-1] –pos1[i]+ gapbetweenchromosomes
tickmarkposition1[chromonumber]=pos1[i]
}
}
tickmarkposition1
plot(pos1,col=chr1)
# This plot shows that the position (pos1) of the SNP markers is increasing. No surprise
here since this is how we defined pos1.It also shows a small vertical gap between markers
at the end of "adjacenct" chromosomes.
The markers have been colored by chromosome (1=black, 2=red, 3=green, 4=blue, etc)
13
# The following function is used to produce Figure 3
# whichchromosomes1=the chromosomes that should be used
#normalize count=divide the number of significant genes by this factor
if (exists("PlotQTLCountplusQTLbars" )) rm(PlotQTLCountplusQTLbars);
PlotQTLCountplusQTLbars =function(NoSignificantGenes, chromosome,LODscore,
MarkerLocation,NormalizationFactorCount=1, whichchromosomes1=c(1:19)
,color1="grey", LocationmQTL) {
NormalizedCount= NoSignificantGenes/ NormalizationFactorCount
rest1=is.element(chromosome, whichchromosomes1)
ylim1=c(0, max( c(NormalizedCount, LODscore )) )
plot(MarkerLocation[rest1], NormalizedCount[rest1],ylim=ylim1, xlab="Mouse
Chromosome",ylab=paste("LOD Score") , cex.axis=1.5,cex.lab=1.2, type="h", xaxt="n",
yaxt="n",col=color1)
text( c(MarkerLocation[rest1])[c( LocationmQTL[rest1])], 0,
labels="*",cex=3,col="red" )
mtext(paste("No.Signif.Genes/", NormalizationFactorCount), side=4,cex=1.5)
for (i in whichchromosomes1) {
rest2=chromosome==i
lines(MarkerLocation[rest2], LODscore[rest2],col="black",lwd=2)
axis(2) # put numerical annotations at the tickmarks in y-axis;
axis(1, tickmarkposition1[whichchromosomes1],
labels=whichchromosomes1,cex.axis=1.5)
} # end of for loop
} # end of function PlotQTLCountplusQTL
# Now we call the function on our count data
par(mfrow=c(2,1),mar=c(4,4,2,1)+.1)
PlotQTLCountplusQTLbars(NoSignificantGenes= NoSignifModuleGenes,
chromosome=chr1,LODscore=lod, MarkerLocation= pos1,
NormalizationFactorCount=20, whichchromosomes1=c(1:10),color1="blue",
LocationmQTL= LocationmQTL )
PlotQTLCountplusQTLbars(NoSignificantGenes= NoSignifModuleGenes,
chromosome=chr1,LODscore=lod, MarkerLocation= pos1,
NormalizationFactorCount=20, whichchromosomes1=c(11:19) ,color1="blue",
LocationmQTL= LocationmQTL)
14
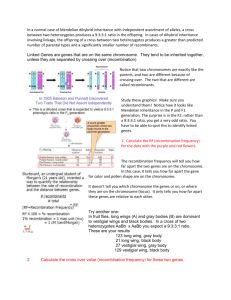
# This is Figure 3 in our article. Blue Module mQTL Profile Partially Overlaps with the
Physiological Trait (Weight) QTL
The black curves represent the QTL analysis across the genome for mouse body weight
using a single marker genome scan according to the LOD score scale to the right.
Significant linkage for weight was found on Chromosome 1 (50.4 Mb, LOD = 4.07),
Chromosome 5 (118.5 Mb, LOD = 3.39), Chromosome 15 (78.2 Mb, LOD = 3.58), and
Chromosome 19 (50.4 Mb, LOD = 4.07). For each marker, the blue vertical line
represents the number (divided by 20) of significant Blue module genes, i.e., the number
of genes whose single-point LOD at this marker is bigger than 2. For example, there is a
SNP marker on Chromosome 2 at which 117 Blue module genes have a LOD bigger than
2. The red stars denote the locations of the mQTLs reported in Table 1.
15
#The following code provides an alternative view of the same curves.
if (exists("PlotQTLCountplusQTL" )) rm(PlotQTLCountplusQTL);
PlotQTLCountplusQTL =function(NoSignificantGenes, chromosome,LODscore,
MarkerLocation,NormalizationFactorCount=1, whichchromosomes1=c(1:19) ) {
NormalizedCount= NoSignificantGenes/ NormalizationFactorCount
rest1=is.element(chromosome, whichchromosomes1)
ylim1=c(0, max( c(NormalizedCount, LODscore )) )
plot(MarkerLocation[rest1],
NormalizedCount[rest1],ylim=ylim1,col=chromosome[rest1], xlab="Marker
Location",ylab=paste("LOD or No.Signif.Genes/", NormalizationFactorCount) ,
cex.axis=1.5,cex.lab=1.2, type="n", xaxt="n", yaxt="n")
text (MarkerLocation[rest1],
NormalizedCount[rest1],label=chromosome[rest1],cex=.75,col=chromosome[rest1])
for (i in whichchromosomes1) {
rest2=chromosome==i
lines(MarkerLocation[rest2], LODscore[rest2],col=chromosome[rest2],lwd=2)
axis(2) # put numerical annotations at the tickmarks in y-axis;
axis(1, tickmarkposition1[whichchromosomes1],
labels=whichchromosomes1,cex.axis=1.5)
} # end of for loop
} # end of function PlotQTLCountplusQTL
par(mfrow=c(2,1),mar=c(3,4,2,1)+.1)
PlotQTLCountplusQTL(NoSignificantGenes= NoSignifModuleGenes,
chromosome=chr1,LODscore=lod, MarkerLocation= pos1,
NormalizationFactorCount=20, whichchromosomes1=c(1:10) )
PlotQTLCountplusQTL(NoSignificantGenes= NoSignifModuleGenes,
chromosome=chr1,LODscore=lod, MarkerLocation= pos1,
NormalizationFactorCount=20, whichchromosomes1=c(11:19) )
16
17
#Recall the LOD score curve for body weight
plot(out.hk,gap=0,col="black")
18
This analysis shows that there are four QTLs with suggestive LOD scores for body
weight on chromosomes 1,5,15,and 19. To see if there are interactions between these four
QTLs one can use the fitqtl function in the qtl package. To know about the details of this
function please visit http://www.biostat.jhsph.edu/~kbroman/qtl/ . In summary, this
function allows one to fit linear models in which the marker locations are used as
independent variables. In our example, we could use the position of the peak markers on
chromosomes 1, 5, 15, and 19 as the independent variables. One could also use
interaction terms between these loci to look for evidence of interaction between the
corresponding QTLs. The output of the fitqtl function will give the p-values associated
with each of the terms used in the model. This allows one to take the insignificant terms
out and rerun the fitqtl function on the model with the significant terms. To run the fitqtl
function on the full model use the following code:
F2crossdata <- sim.geno(F2crossdata, step=2, n.draws=50)
chr<-c(1,5,15,19)
pos<-c(143,118,78,50)
qtl<-makeqtl(F2crossdata,chr,pos)
my.formula<-y~Q1+Q2+Q3+Q4+Q1*Q2+Q1*Q3+Q1*Q4+Q2*Q3+Q2*Q4+Q3*Q4
out.fitqtl <- fitqtl(F2crossdata$pheno[,1], qtl, formula=my.formula)
summary(out.fitqtl)
Summary for fit QTL
Method is: imp
Number of observations:
132
Full model result
---------------------------------Model formula is: y ~ Q1 + Q2 + Q3 + Q4 + Q1 * Q2 + Q1 * Q3 + Q1 * Q4 + Q2 * Q3 +
Model formula is:
Q2 * Q4 + Q3 * Q4
df
SS
MS
LOD
%var Pvalue(Chi2)
Pvalue(F)
Model 32 2592.209 81.00652 22.06625 53.69127 3.578178e-09 6.073403e-07
Error 99 2235.781 22.58364
Total 131 4827.989
Drop one QTL at a time ANOVA table:
---------------------------------df Type III SS
LOD
%var F value Pvalue(Chi2) Pvalue(F)
Chr1@143
14
806.0401
8.8245 16.6951
2.5494
0.000203
0.00368 **
Chr5@118
14
398.7324
4.7039
8.2588
1.2611
0.086
0.24540
Chr15@78
14
813.9587
8.8990 16.8592
2.5744
0.000179
0.00337 **
Chr19@50
14
835.8985
9.1045 17.3136
2.6438
0.000127
0.00263 **
Chr1@143:Chr5@118 4
101.4772
1.2723
2.1019
1.1233
0.210
0.34991
Chr1@143:Chr15@78 4
334.1523
3.9925
6.9211
3.6991
0.001
0.00753 **
Chr1@143:Chr19@50 4
190.4327
2.3430
3.9443
2.1081
0.029
0.08544 .
Chr5@118:Chr15@78 4
25.3292
0.3229
0.5246
0.2804
0.829
0.89005
Chr5@118:Chr19@50 4
116.2888
1.4534
2.4086
1.2873
0.153
0.28019
Chr15@78:Chr19@50 4
118.8034
1.4840
2.4607
1.3151
0.145
0.26963
--Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Note: from this we can conclude that there may be interactions between chromosome 1
and 15 (F statistic p=0.00753) and chromosomes 1 and 19 (F statistic p= 0.08544). Now
19
we go back and instead of using the full model we use the the model which has all the
additive parameters and two interaction terms for chromosomes1 with 15 and 1 with 19.
To do this copy and paste the following code into R:
F2crossdata <- sim.geno(F2crossdata, step=2, n.draws=50)
chr<-c(1,5,15,19)
pos<-c(143,118,78,50)
qtl<-makeqtl(F2crossdata,chr,pos)
my.formula<-y~Q1+Q2+Q3+Q4+Q1*Q3+Q1*Q4
out.fitqtl <- fitqtl(F2crossdata$pheno[,1], qtl, formula=my.formula)
summary(out.fitqtl)
Summary for fit QTL
Method is: imp
Number of observations:
132
Full model result
---------------------------------Model formula is: y ~ Q1 + Q2 + Q3 + Q4 + Q1 * Q3 + Q1 * Q4
df
SS
MS
LOD
%var Pvalue(Chi2)
Pvalue(F)
Model 16 2178.838 136.17738 17.20353 45.12931 2.294350e-10 3.004679e-09
Error 115 2649.151 23.03610
Total 131 4827.989
Drop one QTL at a time ANOVA table:
---------------------------------df Type III SS
LOD
%var F
Chr1@143
10
751.687
7.160 15.569
Chr5@118
2
161.825
1.700
3.352
Chr15@78
6
748.162
7.130 15.496
Chr19@50
6
662.310
6.396 13.718
Chr1@143:Chr15@78 4 374.601 3.791
7.759
Chr1@143:Chr19@50 4 245.341 2.539
5.082
--Signif. codes: 0 '***' 0.001 '**' 0.01 '*'
value Pvalue(Chi2) Pvalue(F)
3.263
2.75e-04 0.000961 ***
3.512
0.020 0.033064 *
5.413
1.13e-05 5.85e-05 ***
4.792
4.99e-05 0.000213 ***
4.065
0.002 0.004046 **
2.663
0.020 0.036070 *
0.05 '.' 0.1 ' ' 1
Note that 45% of the variation in weight phenotype can be explained by the variables of
the model that only has the four loci detected using the scanone function and the two
interaction terms detected by the fitqtl function. If one uses the model for which there are
no interaction terms the model would only explain 34% of the weight variation as can be
seen from the following output
20
F2crossdata <- sim.geno(F2crossdata, step=2, n.draws=50)
chr<-c(1,5,15,19)
pos<-c(143,118,78,50)
qtl<-makeqtl(F2crossdata,chr,pos)
my.formula<-y~Q1+Q2+Q3+Q4
out.fitqtl <- fitqtl(F2crossdata$pheno[,1], qtl, formula=my.formula)
summary(out.fitqtl)
Summary for fit QTL
Method is: imp
Number of observations:
132
Full model result
---------------------------------Model formula is: y ~ Q1 + Q2 + Q3 + Q4
df
SS
MS
LOD
%var Pvalue(Chi2)
Pvalue(F)
Model
8 1664.924 208.11546 12.12143 34.48483 3.056179e-09 9.729804e-09
Error 123 3163.066 25.71598
Total 131 4827.989
Drop one QTL at a time ANOVA table:
---------------------------------df Type III SS
LOD
%var F value Pvalue(Chi2) Pvalue(F)
Chr1@143 2
224.329
1.964
4.646
4.362
0.011
0.01479 *
Chr5@118 2
292.051
2.531
6.049
5.678
0.003
0.00438 **
Chr15@78 2
339.305
2.921
7.028
6.597
0.001
0.00190 **
Chr19@50 2
339.843
2.925
7.039
6.608
0.001
0.00188 **
THE END
21