Supplementary Methods - Word file
advertisement

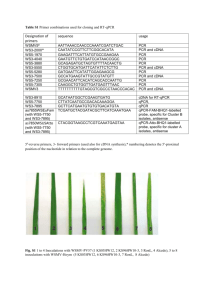
Supplementary Methods S5. cDNA isolation Sequencing of a cDNA isolated from a Lotus root/nodule primordia library by PCR amplification reveal a transcription start site at least 30 nucleotides upstream of the start codon and a 3’ untranslated region of 188 nucleotides. Alignment of genomic and cDNA sequences defined a primary structure of CCaMK consisting of 7 exons. Double mutant analysis Homozygous nfr1-1snf1-1 double mutants were identified in a segregating population. F2 progeny was screened in absence of M. loti and DNA extracted from spontaneously nodulating plants. Homozygous nfr1-1 snf1-1 double mutants were identified by sequencing of PCR fragments encompassing their mutations. In the next generation a total of more than hundred plants originating from selfing of two different nfr1-1snf1-1 double mutants were confirmed to develop spontaneous nodules and PCR fragments of 15 plants from each progeny were sequenced to confirm presence of homozygous nfr1-1 and snf1-1 mutations. PCR fragments encompassing nfr1-1 and snf1-1 mutant alleles respectively, were amplified using the following primers: K3-rev1-fw1 5`TAAACACCTATCGCATAAGTG3` 16K18rev-rev2 5`CAGCAACTAATTCACCTGTC3` Ca2Fwd 5`TGGCTTGCATCCAAACGGC3` Ca3-3Rev 5`ACTATTGTTGTCTCACTTTAGTG3` The har1-1 snf1-1 double mutants were identified using the same crossing strategy and scoring for hypernodulation. In F3, ten plants developing an excess number of spontaneous nodules were confirmed to be har1-1 snf1-1 double mutants by sequencing the snf1-1 allele and by PCR amplification of the deletion allele present in har1-1. Presence of har1-1 was confirmed using the following primers: har1 #3fw2 5`CCTGAAATGCCTATTCGTTGAG3` #3rev2 5`CACAGCTTCTTCTGCATGCG3` Nin-GUS reporter lines The Nin-GUS snf1-1 lines were identified in an F2 population of 475 grown for 10 days on B & D media2. 125 plants were inoculated with 100 times diluted 48 hours culture of M. loti or M. loti R7A NodC, respectively, and 125 plants were left uninoculated. For each treatment presence of Nin-GUS in five snf1-1 homozygous plants was confirmed by sequencing PCR fragments encompassing the snf1-1 mutation. Presence of Nin-GUS was shown by amplification of a fragment spanning part of the Nin promoter and GUS coding region using the following primers. Pro6+ 5´GAGAGGCAGCAAGCATATAC3` Gusrev1 5´CGCGATCCAGACTGAATGCCC3` Expression Analysis. Using a RNeasy Plant Mini kit and RNase-free DNase kit (Qiagen), total RNAs were prepared from various organs of twenty days old plants inoculated with MAFF30–3099. The first strand of cDNA was synthesized with Omniscript RT kit (Qiagen). Quantitative PCR was performed on an ABI PRISM® 7000 Sequence Detection System (Applied Biosystems) using QuantiTect™ SYBR® green PCR kit (Qiagen) to amplify the target transcripts from diluted cDNA. Sample volumes were normalized for equal amplification of DNA fragments with primers specific for Actin (BU494092). PCR cycling conditions comprised an initial denaturation step at 95° C for 15 min followed by 40 to 45 cycles at 94° C for 15 s, 52° C for 30 s, and 72° C for 45 s. The primers used for transcript amplification were: LjActin-F, 5’- TGTGAGTCACACTGTGCCAATC -3’; LjActin-R, 5’- GACCAGCAAGATCCAAACGAA -3’; LjCCaMK-F, 5’- GGA GAC AAT GCA ACT CTG TCT GA -3’; (1153bp-1230bp: 374aa-400aa: GDNATLSEFVEVLKAMKMPSLIPLAP) LjCCaMK-R, 5’- CGG TGC TAG AGG GAT CAA TGA G -3’; LjNin1-F, 5’- TTC TCT GCT TTT CTT GAG CTT T -3’; LjNin1-R, 5’- TTG GGA TGG GAT TGG GAT TG -3’; Melting curve analysis was used to determine PCR product homogeneity. Actin (BU494092) was used as calibrator. Amplified sequences was confirmed by sequencing. Results in Figure 2 are shown as fold increase compared to leaves in (b) and uninoculated roots (c and d at time zero). Actin was used as internal control and relative values normalised to the uninoculated root (zero days) are shown. In vitro kinase assays. The CCaMK cDNA encoding region was amplified by PCR with primers 5’-CACCATGGGATATGATCAAACCAG-3’ and 5’-AAACTATGATGGACGAAGAG-3’. Point mutations were inserted by PCR-based site-directed mutagenesis using the following primer sets; G30E, 5’-GTCAGAAAAGAAACCAAAAAATCAGG-3’ and 5’-CCTGATTTTTTGGTTTCTTTTCTGAC-3’; T265I, 5’-CTATGAGAAGATTTGGAAGGGC-3’ and 5’-GCCCTTCCAAATCTTCTCATAG-3’. PCR products were cloned into pENTR/SD/D/TOPO vector (Invitrogen) and resulting entry clones were confirmed by sequencing for carrying targeted mutations and no PCR error. To make maltose-binding protein (MBP)-tagged CCaMK variants, insert sequences of the entry clones were converted into the pKM596 vector (Fox, J. D. & Waugh, D. S. Maltose-binding protein as a solubility enhancer. Methods Mol Biol. 2003, 205, 99-117) by Gateway LR reaction (Invitrogen). Expression of MBP-tagged CCaMK and its mutant variants was induced by adding 0.5 mM IPTG and cultured at 28˚C for 2 hrs in E. coli strain Rosetta pLaqI (Novagen). Protein purification using amylose resin (New England Biolabs) was performed according to manufacturer’s instruction. Purified proteins were desalted on a PD-10 column (Amersham) and eluted with 50 mM HEPES (pH7.4) buffer. Protein amounts were determined by the Bradford method (Biorad) using BSA as a standard, and adjusted according to Coomassie staining visualisation in a SDS-PAGE gel. In vitro kinase assay was carried out using 0.4 µM CCaMK protein per assay (Fig4c, d) in total 25 µl reaction for 30 min (Fig.4c) or 5 min (Fig.4 d,e) at 25˚C in presence of 50 mM HEPES (pH7.4) containing 10 mM (Fig4c) or 5 mM (Fig4d, e) magnesium chloride, 1 mM dithiothreitol, 200 µM ATP and 10 µCi [-32P] ATP. Either 4 mM EGTA or 0.1 mM calcium chloride with or without 1 µM bovine brain calmodulin (Sigma) were added as indicated. Five µg myelin basic protein (Sigma) was used as substrate. Kinase reactions were stopped by adding SDS-PAGE sample buffer and boiling for 5 min. Samples were separated by SDS-PAGE electrophoresis and the gel was subsequently stained by Coomassie brilliant blue R-250. Radioactive bands were visualised by phosphorimager (Molecular Dynamics) and quantified using Image Quant (version 5.2, Molecular Dynamics) software. Representative data from at least three repetitions are shown in Fig. 4. The trendlines for data in Fig. 4e are following the equation; y = 6.8738x2 + 0.0671x (R2 = 0.9918) see also Fig. S4.