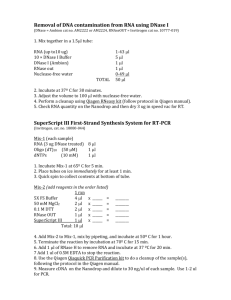
RNA Protocols Table of Contents

RNA Protocols Table of Contents
Last revised: 1/10/2012 By: Tim Starr
RNA Protocols Table of Contents ......................................................................................................... 1
Qiagen Rneasy Mini Kit ........................................................................................................................ 2
Qiagen RNeasy Midi kit: ....................................................................................................................... 4
Invitrogen Purelink Total RNA Purification System ............................................................................... 6
DNase I Treatment: Deoxyribonuclease I, Amplification Grade (Invitrogen) ........................................ 8
Trizol RNA isolation ............................................................................................................................ 10
Dnase treat the RNA using Ambion Turbo DNA-Free kit .................................................................... 11 cDNA synthesis using Invitrogen's VILO kit ........................................................................................ 12
RNA Extraction from FFPE Block Slide Tissue ................................................................................... 13
RNA T. Starr Page 1 4/13/2020
Qiagen Rneasy Mini Kit
Background: Isolate RNA from single cell suspension using Qiagen RNeasy micro or mini kits. Remember, RNase is everywhere. All steps are done at room temperature. Working faster will increase RNA yield, but will also increase your mistakes.
Procedure using Qiagen RNeasy Mini kit:
* Collect cells in RP10, count cells, remove any aliquots for FACs analysis. Note: Use mini kit if you have between 1
– 10 e6 cells. Otherwise use the midi or maxi kits. One RNeasy column can only bind 100 µg RNA and will only take 1e6 cells. Using more cells will require more columns.
* Spray preparattion area with RNase AWAY. Respray area continuously throughout the procedure if you touch areas outside the RNA isolation area.
* Make up fresh RLT/
-ME buffer first – (10 l
-ME/1 ml Buffer RLT) (good stored at RT for up to 1 month)
* Spin cells, remove as much liquid as possible, loosen pellet by vortexing or flicking tube before adding lysis buffer
* Resuspend in Buffer RLT/
ME (600ul/10e6 cells or 350µl/5e6 cells or less). Buffer volume = ______
Lysis of cells: Lysis can be accomplished in one of three ways: Homgenization, Syringe, or Qiashredder columns. Can also first do syringe and then qiashredder columns.
Syringe
* Lyse through syringe (18G1) 5-10 times, be thorough & try not to touch the cell samples at this point.
Qiashredder columns
* Aliquot a maximum of 600
l through Qiashred columns (spin for 2 minutes at 14,000 rpm). # of columns = ______.
* Throw away columns, RNA at this point is in the liquid.
Homogenizer
* Insert homogenizer and turn on for 15-90 seconds. Homogenizer has a 7mm diameter blade, use #2 setting. Keep foaming to a minimum by using properly sized tubes and keep the tip submerged and hold tip to one side of tube. If sample is in 500 µl or less use a microcentrifuge tube, otherwise use either a FACs tube or a 15 ml conical tube.
* Wash homogenizer by running first in RNase-free EtOH and then run in DEPC-treated water between each sample.
After lysis continue as follows:
* Add an equal volume of 70%EtOH to lysate, mix well by pipetting
* Add up to 700 µl lysate to an RNeasy column, spin 15 seconds at 10,000 rpm, empty column waste and replace tube under column, can add another 700 µl to same tube a spin again, or just continue.
* RNA at this point is stuck in the white pads of the columns.
* Add 350ul Buffer RW1, spin 15 seconds at 10,000 rpm. Throw away waste from tubes and put tubes back under columns.
* Mix enough DNase with RDD buffer (10 µl DNase to 70 µl RDD buffer) to treat columns with 80 µl. Add 80 µl
RDD/DNase buffer to columns and incubate 15 min RT.
* Add an additional 350 µl RW1 buffer, spin 15 seconds 10,000 rpm. This step removes any remaining DNA in the columns.
* Transfer columns to clean 2 ml collection tubes, add 500ul RPE buffer, spin 15 seconds at 10,000 rpm.
RNA T. Starr Page 2 4/13/2020
* Carefully transfer columns to clean 2 ml collection tubes, being careful not to let columns touch liquid in tube. Add another 500 µl RPE buffer, spin 2 minutes at 10,000 rpm. This step should remove the ethanol from the previous step.
* Transfer columns to clean 1.5 ml collection tubes. Add 30 to 50 µl H
2
O, let sit 1 minute, spin 1 min 10,000 rpm. If expected RNA content is > 30 µg repeat elution by either adding another 50 µl H
2
O or by re-adding the first eluate and spin 1 min 10,000 rpm. Re-adding the first eluate will reduce RNA yield by 15 – 30%, but it will increase RNA concentration.
* At this point the RNA is in the water, and has been washed from the columns.
* Combine eluted RNA into one tube. Final Volume = ______
* Determine the amount of RNA you collected by measuring OD using spectrophotometer or using ribogreen plates.
(see separate protocol)
* Freeze @ 70ºC (I usually store at -20°C).
Qiagen Rneasy Mini kit:
Maximum binding capacity 100 ug RNA
Maximum loading volume
RNA size distribution
Minimum elution volume 30 ul
Maximum amount of starting material 10e7 animal cells
Average yield of total RNA (ug):
700 ul
RNA >200 nucleotides
Cell cultures (10e6)
Mouse tissue: (10 mg)
Brain
Heart
10-35
8
10
Kidney
Liver
Spleen
35
40
35
Thymus 45
Lung 10
Recommended to use 3-4x10^6 cells if RNA content of cells is unknown. (Overloading the column will significantly reduce yield and purity)
All steps should be performed at room temperature, and work quickly.
RNA T. Starr Page 3 4/13/2020
Qiagen RNeasy Midi kit
:
* Collect cells in RP10, count cells, remove any aliquots for FACs analysis. Note: Use midi kit if you have between
5e6 – 1e8 cells. Otherwise use the midi or maxi kits. One RNeasy column can only bind 1.0 mg RNA and will only take
1e8 cells.
* Spray preparattion area with RNase AWAY. Respray area continuously throughout the procedure if you touch areas outside the RNA isolation area.
* Make up fresh RLT/
-ME buffer first – (10 l
-ME/1 ml Buffer RLT) (good stored at RT for up to 1 month). (If you have between 5e7 cells to 1e8 cells use 4 ml of buffer/sample: for 5e6 to 3e7 cells use 2 ml buffer/sample)
* Spin cells, remove as much liquid as possible, loosen pellet by vortexing or flicking tube before adding lysis buffer
* Resuspend in Buffer RLT/
-ME (2 ml/5e6 to 3e7 cells or 4 ml/5e7 to 1e8 cells. In between those two amounts, its your judgement call). Buffer volume used in this experiment = ______
* Lyse cells by either using a homogenizer or a syringe and needle.
Homogenizer
* Insert homogenizer and turn on for 45 seconds. Homogenizer has a 7mm diameter blade, use #2 setting. Keep foaming to a minimum by using properly sized tubes and keep the tip submerged and hold tip to one side of tube. Use a
FACs tube or a 15 ml conical tube.
* Wash homogenizer by running first in RNase-free EtOH and then run in DEPC-treated water between each sample.
Syringe
* Lyse through syringe (18G RNase-free needles) 5-10 times, be thorough & try not to touch the cell samples at this point.
After lysis continue as follows:
* Add an equal volume of 70%EtOH to lysate, mix well by shaking, make sure there is no precipitate (do not centrifuge).
* Add up to 4 ml lysate to an RNeasy midi column held in a 15 ml conical tube, spin 5 min at 3-5,000 rpm, empty column waste and replace tube under column, can add another 4 ml to same tube a spin again, or just continue. RNA at this point is stuck in the white pads of the midi column.
* Add 2 ml Buffer RW1, spin 5 min at 3-5,000 rpm. Throw away waste from tubes and put tubes back under columns.
* Mix enough DNase with RDD buffer (20 µl DNase to 140 µl RDD buffer) to treat columns with 160 µl. Add 160 µl
RDD/DNase buffer to columns and incubate 15 min RT. Make sure DNase is pipetted onto the silica-membrane gel and not the sides of the columns.
* Add an additional 2 ml RW1 buffer, let sit for 5 min then spin 5 min 3-5,000 rpm. This step removes any remaining
DNA in the columns. Discard the flow-thru, reuse the collection tube. RNA is still in the silica-gel at this point.
* Add 2.5 ml RPE buffer to columns, spin 2 min at 3-5,000 rpm. (Make sure ethanol has been added to the RPE buffer). Discard flow thru.
* Add another 2.5 ml RPE buffer and spin 5 min at 3-5,000 rpm. Make sure no ethanol is allowed back into the column when removing the column from the tube.
* Carefully transfer columns to clean 15 ml collection tubes, being careful not to let columns touch liquid in tube. Add
150 to 250 µl H
2
O (add only 150 if expecting less than 150µg RNA), let sit 1 minute, spin 3 min at 3-5,000. Repeat by adding another 150 - 250 µl H
2
O or by re-adding the first eluate and spin 3 min at 3-5,000 rpm. Re-adding the first eluate will reduce RNA yield by 15 – 30%, but it will increase RNA concentration.
* At this point the RNA is in the water, and has been washed from the columns.
RNA T. Starr Page 4 4/13/2020
* Combine eluted RNA from multiple samples into one tube. Final Volume = ______
* Determine the amount of RNA you collected by measuring OD using spectrophotometer or using ribogreen plates.
(see separate protocol)
* Freeze @ 70ºC (I usually store at -20°C).
Qiagen Rneasy Midi kit:
Maximum binding capacity
Maximum loading volume
RNA size distribution
Minimum elution volume
1.0 mg RNA
4 ml
RNA >200 nucleotides
300 ul
Maximum amount of starting material 10e8 animal cells
Average yield of total RNA (ug):
Cell cultures (Hela) (7.0e7) 1000
Mouse tissue: (100 mg)
Brain
Heart
Kidney
100
200
200
Liver
Spleen
Thymus
200
200
100
Lung 100
Recommended to use 3-4x10^7 cells if RNA content of cells is unknown. (Overloading the column will significantly reduce yield and purity)
All steps should be performed at room temperature, and work quickly.
RNA T. Starr Page 5 4/13/2020
Invitrogen Purelink Total RNA Purification System
Fresh or frozen animal tissue
Amount of tissue
≤10 mg
10 –60 mg
60 –100 mg
100 –200 mg
RNA Lysis Solution (ml)
0.3*
0.6
1.2
2.4
Place tissue in 5 ml FACs tube on ice.
2mercaptoethanol (µl)
3
6
12
24
Add appropriate amount of RNA lysis solution and 2-mercaptoethanol
Homogenize using a rotor stator for 45 seconds at high speed. Wash rotor-stator three times in between samples
Centrifuge at ~2,600 × g for 5 minutes at room temperature.
Carefully transfer the supernatant to a clean RNase-free tube.
Add one volume of 70% ethanol to each volume of tissue homogenate.
Mix thoroughly by shaking or vortexing. Disperse any visible precipitate that may form after adding ethanol.
Transfer up to 700 μl of the sample (including any remaining precipitate) to the RNA Spin Cartridge pre-inserted in a collection tube.
Centrifuge at 12,000 × g for 15 seconds at room temperature. Discard the flow-through, and re-insert the cartridge in the tube.
Repeat Steps 3 –4 until the entire sample has been processed.
Optional: If your downstream application requires DNA-free total RNA, you may use the convenient on-column DNase I treatment at this point in the procedure (see page 38).
After loading the sample onto the spin cartridge, perform the following DNase I digestion procedure before proceeding to the following wash step
Add 350 μl of Wash Buffer I to the spin cartridge.
Centrifuge at 12,000 × g for 15 seconds at room temperature. Discard the flow-through and the collection tube.
Add the following items supplied with DNase I, Amplification Grade, to a clean, RNase-free microcentrifuge tube:
Component
DNase Buffer
DNase I (1 unit/μl)
Final Volume
Volume
70 μl
10 μl
80 μl
Mix gently by inverting the tube. Centrifuge briefly to collect the contents of the tube.
Transfer the solution to the center of the spin cartridge, and incubate for 15 minutes at room temperature.
Add 350 μl of Wash Buffer I to the spin cartridge. Centrifuge at 12,000 × g for 15 seconds at room temperature.
Discard the flow-through.
Proceed with the following wash step
Add 700 μl of Wash Buffer I to the spin cartridge. Centrifuge at 12,000 × g for 15 seconds at room temperature. Discard the flow-through and the collection tube.
Place the spin cartridge into a clean RNA Wash Tube, provided in the kit.
Add 500 μl Wash Buffer II with ethanol (prepared as described on page 7) to the spin cartridge.
Centrifuge at 12,000 × g for 15 seconds at room temperature. Discard the flow-through, and re-insert the cartridge in the tube.
RNA T. Starr Page 6 4/13/2020
Repeat Steps 8 –9 once.
Centrifuge the spin cartridge at 1 2,000 × g for 1 minute at room temperature to dry the membrane with attached RNA.
Discard the collection tube, and insert the cartridge into an RNA Recovery Tube supplied with the kit.
To elute the RNA, add 30 –100 μl of RNase-free water to the center of the spin cartridge, and incubate at room temperature for 1 minute. See page 7 for detailed elution parameters.
Centrifuge the spin cartridge for 2 minutes at ≥12,000 × g at room temperature. If you are performing sequential elutions, collect all elutes into the same tube.
To determine the quantity and quality of the RNA, see page 30.
To perform DNase I digestion of the purified RNA, see page 39.
If you will use the RNA within a few hours, store on ice. For longer storage, store at –80°C.
RNA T. Starr Page 7 4/13/2020
DNase I Treatment: Deoxyribonuclease I, Amplification Grade (Invitrogen)
Description:
Deoxyribonuclease I, Amplification Grade (DNase I, Amp Grade) digests single-and double-stranded DNA to oligodeoxy-ribonucleotides containing a 5′- phosphate. It is suitable for eliminating DNA during critical RNA purification procedures such as those prior to RNA-PCR amplification. DNase I, Amp Grade is purified from bovine pancreas and has a specific activity of ≥ 10,000 U/mg.
Components:
18068-015 DNase I, Amp Grade
Y02340 10X DNase I Reaction Buffer
Y02353 25 mM EDTA (pH 8.0)
Unit Definition:
One unit increases the absorbance of a high molecular weight DNA solution at a rate of 0.001 A260 units/min/ml of reaction mixture at 25ÅãC.
Storage Buffer: Unit Assay Conditions:
20 mM sodium acetate (pH 6.5) 0.1 M sodium acetate (pH 5.0)
5 mM CaCl2 5 mM MgCl2
0.1 mM PMSF 50 É g/ml calf thymus DNA
50% (v/v) Glycerol
10X DNase I reaction Buffer:
200 mM Tris-HCl (pH 8.4), 20 mM MgCl2, 500 mM KCl
Quality Control:
The ability to digest double-stranded DNA to oligonucleotides is confirmed. DNase I and components are tested for the absence of RNase activity by incubating with 0.24-9.5 Kb RNA Ladder; gel analysis shows no degradation of the RNA ladder as compared to a no-DNase I control. The enclosed buffers were assayed with the enzyme and met quality control specifications.
Protocols:
Preparation of RNA Sample Prior to RT-PCR :
Prepare duplicate tubes if positive and negative reverse transcriptase (RNA) samples are to be used in the amplification reaction. Add the following to an RNase-free, 0.5-ml microcentrifuge tube on ice:
1 µg RNA sample
1 µl 10X DNase I Reaction Buffer
1 µl DNase I, Amp Grade, 1 U/µl
DEPC-treated water to 10 µl
NOTE: To work with larger amounts of RNA, scale up the reaction (including volume) linearly. Incubate tube(s) for 15 min at room temperature. Inactivate the DNase I by the addition of 1
µl of 25 mM EDTA solution to the reaction mixture. Heat for
10 min at 65C. The RNA sample is ready to use in reverse transcription, prior to amplification.
NOTE: It is important not to exceed the 15-minute incubation time or the roomtemperature incubation. Higher temperatures and longer time could lead to Mg++ dependent hydrolysis of the RNA. Additionally, it is vital that the EDTA be added to at least 2 mM prior to heat-inactivation to avoid this problem.
RNA T. Starr Page 8 4/13/2020
Another option is the Ambion Turbo DNA-fre Kit
RNA T. Starr Page 9 4/13/2020
Trizol RNA isolation
To isolate RNA from plated cells (NIH 3T3, eg).
* Aspirate media from plated cells.
* Add 1 ml Trizol per 10 cm 2 of plate (independent of confluence)
6 well plate = 35 mm plate = 1 ml trizol
24 well plate = 15 mm plate = 1.77 cm 2 = 177 µl trizol (I’ll use 200 µl per well)
* Mix extensively and transfer to 1.5 ml eppe.
Note: If there is a lot of insoluble material after a 5 minute RT incubation, can remove by centrifugation at 12,000 g for 10 min at 4º C before adding chloroform (a clear supernatant and jelly-like pellet should be seen). Remove supernatant and proceed to next step. Only do this if you only want RNA, as it will destroy the DNA. There is an alternate procedure if you are also collecting DNA (see Trizol protocol from manufacturer).
* Optional: Can add glycogen to the sample which may improve yield. It will remain with RNA as it is soluble in water.
* Incubate 5 minutes RT. Add 200 µl chloroform (no isoamyl alcohol is needed) for every 1 ml of
TRIzol. Shake vigorously for 15 seconds and incubate at room temp for 2-3 min. Centrifuge samples 15 min at 12,000 g at 4º C. There should be good phase separation at this point.
* Transfer upper (aqueous) phase to a clean tube. Save the lower phase for later protein extraction.
* Add either 0.75 ml of ethanol per ml of TRIzol used initially or 0.5 ml of isopropanol per ml of TRIzol used. Incubate at room temp for 10 min (if cloudy, precipitate for an additional 10
– 15 min).
Centrifuge 12,000 g 10 min at 4ºC. RNA should be visible on side of tube at this point. Can store at 4ºC overnight at this point (do not store at -20ºC and don’t go longer than overnight).
* Remove supernatant. Wash pellet with 1 ml 75% ehtanol for every 1 ml of TRIzol used. Mix by flicking and inverting tube or vortexing and centrifuge at 7,500 g 5 min at 4º C. Can store in
75% ethanol for 1 week at 4ºC or 1 year at -20ºC.
* Remove ethanol and air dry (preferable to vacuum drying). Pellet should be white or clear jelly-like.
If pellet is overdried, can resuspend in Rnase free H2O and m ay have to be incubated at 55ºC for 10-15 min with repeated pipetting to completely dissolve. In general redissolve RNA from
5e6 HeLa cells in 50 µl of DEPC water.
* If using RNA for RT-PCR, treat with Dnase I (see Dnase protocol)
Expected yield from
1e6 cells = 1 to 15 µg depending upon cell type
1e4 cells = 140 ng (using glycogen)
10 mg tissue = 20 to 77 µg depending upon tissue type
Expected A260/A280 ratio: Dissolved in H2O = 1.785, dissolved in TE = 2.2. Note: If ratio is < 1.65 see detailed manual for improving.
RNA T. Starr Page 10 4/13/2020
Dnase treat the RNA using Ambion Turbo DNA-Free kit
* Two different methods, depending upon whether you have normal or severe DNA contamination.
(Severe can occur in certain tissues like spleen, kidney and thymus). The following protocol is for normal DNA contamination (up to 50 µg/ml RNA).
* Normally done in a 50 µl reaction (10 to 100 µl) with up to 10 µg of RNA
* Add 0.1 volume 10X turbo Dnase buffer and 1 µl Turbo Dnas to the RNA and mix gently. Incubate
37ºC for 20-30 min.
* Add 0.1 volume (or 2 µl, whichever is greater) resuspended (vortexed) Dnase Inactivation Reagent and mix well. Incubate 2-3 minutes room temp, mixing three times during incubation.
*Centrifuge 10,000 g for 1.5 min. transfer RNA to a clean tube.
RNA T. Starr Page 11 4/13/2020
cDNA synthesis using Invitrogen's VILO kit
4 µl
2 µl x µl to 20 µl
5x VILO Reaction Mix
10X SuperScript Enzyme Mix
RNA (up to 2.5 µg)
H2O
Can scale up to 100 µl
Gently mix tube, incubate 25º C 10 min incubate 42ºC 60 min incubate 85ºC 5 min (to terminate reaction).
.
Dilute 1:20 if starting with 2 or more µg RNA
Store at 20ºC.
RNA T. Starr Page 12 4/13/2020
RNA Extraction from FFPE Block Slide Tissue
From Luke Manlove
Incubate 20 min in xylenes at 37C
Scrape off stuffs into a 1.5mL eppendorf tube using a razor blade
Be super careful to get all the tissue and try not to ding it up too much
Spin down @ 15000rpm for 10min at 2C
Decant the xylenes
Add fresh xylenes
Spin at 15000rpm 10min 2C
Wash w/ 500uL EtOH 100%
Spin at 15000rpm for 10min at 2C
Remove EtOH completely
Add 720uL tail buffer + 80uL Proteinase K
Tail buffer should have ~1mol/L guanidinium thiocyanate, 25mmol/L 2 mercaptoethanol, SDS, and
Tris w/ pH~7.5
Shake O/N at 55C
Add 80uL ProK
Shake O/N 55C
Add 80uL ProK
Shake O/N 55C
Extract w/ acid phenol:CHCl3
Ph: CHCl3 70:30, pH~4.3
Spin 10 min 15000rpm 2C
Remove aqueous phase and put in fresh eppendorf tube
Add 500uL 2-propanol
Incubate -20C for 30min
Spin for 30min at 14000rpm
Decant isopropanol
Wash pellet in 70% EtOH
Resuspend in H2O (RNase-free!) (20uL)
Add 500uL Trizol
Incubate and invert, etc to mix in the Trizol
Spin at 12000x g for 10min at 2C
Extract aqueous layer to a new eppendorf tube
Add another aliquot of 500uL Trizol
Spin at 12000x g for 10min at 2C
Move aqueous (top) phase to a new tube
Add 0.5mL Isopropyl-OH/1.0mL Trizol (0.5mL Isopropyl-OH/5x106cells)
Incubate at room temp for 10 min
Spin at 12,000xg for 10 min at 2-8C
Remove the supernatant
Wash the RNA pellet w/ 1mL 75%Et-OH/5x106cells
Vortex the sample to mix
Spin at 7500xg for 5min at 2-8C
Air-dry the pellet for 5-10min
Dissolve RNA in 50uL RNAse-Free H2O by pump-pipetting
Incubate at 55-60C for 10min
A260/280 ratio should be at least 1.65, lower than this is indicative of poor dissolving or protein contamination
RNA T. Starr Page 13 4/13/2020
RNA T. Starr Page 14 4/13/2020