Four ml cuvette version
advertisement

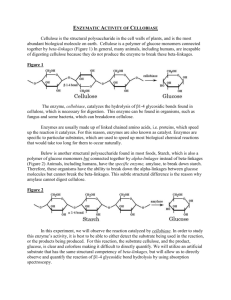
ENZYMATIC ACTIVITY OF CELLOBIASE Cellulose is the structural polysaccharide in the cell walls of plants, and is the most abundant biological molecule on earth. Cellulose is a polymer of glucose with beta linkages connecting the glucose molecules (figure below). In general, animals, including humans, are incapable of digesting cellulose, because they do not produce the enzyme to break these beta-linkages. Enzymes are usually proteins that speed up the reactions (catalyst) that would take too long without the enzyme. Enzymes speed up most biological chemical reactions. Animals have the enzyme to break down starch, and therefore they have the ability to break the alpha-linkages between glucose molecules but animals cannot break the beta-linkages and therefore they cannot digest cellulose. The enzyme, cellobiase, catalyzes the hydrolysis of β1-4 glycosidic bonds found in cellulose. This enzyme can be found in organisms, such as fungus and some bacteria, which can breakdown cellulose. In order to study an enzyme it is best to be able to either detect the substrate being used up by the reaction, or the products being made. Both the substrate (cellulose) and the products glucose, for the reaction catalyzed by cellobiase are clear/colorless, and therefore difficult to detect quantitatively. Extraction of Cellobiase Enzyme 1. 2. 3. Weigh 4 grams of mushroom and grind with 8 ml of extraction buffer with a mortar and pestle until you have a slurry. Filter slurry through a coffee filter into (3) 2 ml microcentrifuge tubes. Spin for 2 mins to pellet debris. Transfer the supernatant (liquid) to a clean 15 ml tube. Make sure to not transfer any solid material. This is your enzyme extract. Measuring Enzymatic Activity of Cellobiase To make this reaction easier to follow, an artificial substrate, p-nitrophenyl glucopyranoside, will be used. This artificial substrate can also bind to the enzyme and be broken down similarly to cellulose. When the artificial substrate, p-nitrophenyl glucopyranoside, is broken down by cellobiase, it produces glucose and p-nitrophenol. When p-nitrophenol is mixed with a basic solution referred to as the stop solution, it will stop the reaction and turn the solution yellow. The amount of yellow color is proportional to the amount of p-nitrophenol present. For every molecule of p-nitrophenol present, one molecule of p-nitrophenyl glucopyranoside has been reacted with by cellobiase. For the cellobiase reactions being run, another advantage of using a basic solution to develop the color of the p-nitrophenol is that the basic pH will also denature the enzyme and stop the reaction. Substrate p-nitrophenyl--D glucopyranoside Colorless Solution Products Glucose p-Nitrophenol Yellow in Stop Solution The yellow color of p-Nitrophenol can be measured using a spectrophotometer. Therefore, more p-Nitrophenol correlates to an increase in color intensity in a linear relationship. Determining Cellobiase Enzymatic Activity 1. Label (2) 15 ml centrifuge tubes, one labeled Enzyme the other labeled Control. Add 9 ml of 1.5 mM p-nitrophenyl--D glucopyranoside substrate to each. 2. Next, label 7 cuvettes #1-7 (these will be your time points, see table below), then pipet 2 ml of Stop Buffer into each cuvette. What is the purpose of the Stop Buffer? Next, you will be adding your enzyme extract to the 15 ml tube labeled Enzyme and immediately removing 2 ml of this mixture then adding it to cuvette labeled #1, this is be your zero time point. It is important that at this same time that you need to start a timer for the remaining time points, (see table below). 3. Add 4.5 ml of your enzyme extract to the 15 ml tube labeled Enzyme, mix and immediately remove 2 ml of this and add it to cuvette labeled #1, and mix the solution in the cuvette. 4. After 1 min, remove an additional 2 ml of your reaction from the 15 ml tube labeled Enzyme and add it to cuvette labeled #2 and mix briefly. 5. Remove 2 ml of your reaction at 2 mins and add it to cuvette labeled #3. 6. Remove 2 ml of your reaction at 4 mins and add it to cuvette labeled #4. 7. Remove 2 ml of your reaction at 6 mins and add it to cuvette labeled #5. 8. Remove 2 ml of your reaction at 8 mins and add it to cuvette labeled #6. 9. Add 2 ml of the control and add it to cuvette labeled #7, this should be your blank. You will measure the absorption of the p-Nitrophenol generated in the reaction with the enzyme extract after you determine a standard curve for p-Nitrophenol. Cuvette 1 2 3 4 5 6 7 Time 0 min 1 min 2 min 4 min 6 min 8 min Control Absorption p-Nitrophenol Concentration Determining Standard Curve of p-Nitrophenol The concentration of p-Nitrophenol can be determined by comparing to a standard curve of known concentrations of p-Nitrophenol. You will make this standard curve using p-Nitrophenol ranging from 12.5 nM to 100nM p-Nitrophenol. When a graph of absorbance vs. concentration is plotted for the standard solutions, a direct relationship should result. The direct relationship between absorbance and concentration for a solution is known as Beer’s law. You can determine the p-nitrophenol concentration of your prepared sample by measuring its absorbance and then finding the corresponding concentration on the standard curve. Because the relationship is linear, you could also calculate the p-nitrophenol concentration using the formula for the standard curve. 1. For the p-Nitrophenol standards, label 5 cuvettes with the known concentrations of the p-Nitrophenol standards (see the data table). 2. Add 2 ml of water to cuvette 1, 1.75 ml of water to cuvette 2, 1.5 ml of water to cuvette labeled #3, 1 ml to cuvette labeled #4 and no water to cuvette labeled #5. 3. Add no p-Nitrophenol to cuvette 1, 0.25 ml of p-Nitrophenol to cuvette labeled #2, 0.5 ml p-Nitrophenol to cuvette labeled #3, 1 ml of p-Nitrophenol to cuvette labeled #4 and 2 ml of p-Nitrophenol to cuvette labeled #5. 4. Add 2 ml Stop Buffer into each cuvette, mix by vortex or invert several times immediately. 5. Read the absorbance at wavelength of 410 nm using cuvette labeled #1 as your blank. Setting up your Standards: Cuvette 1 2 3 4 5 p-Nitrophenol concentration 0 (blank) 12.5 nM 25.0 nM 50.0 nM 100 nM 0.2 mM p-Nitrophenol 0.00 ml 0.25 ml 0.50 ml 1.00 ml 2.00 ml Water Stop Buffer 2.00 ml 1.75 ml 1.50 ml 1.00 ml 2.00 ml 2.00 ml 2.00 ml 2.00 ml 2.00 ml 0.00 ml Absorption @ 410 nm 0 Use Excel to find out the equation for the linear best fit line and write it below (these instructions are for Excel in Windows 7, yours may differ): 1. Open an Excel spread sheet containing a column for p-Nitrophenol standard concentrations, and an additional column with their corresponding absorbances (as shown above). 2. Highlight the values, click on insert, and choose the scatter plot with no line function. 3. Go to chart layouts, choose trendline. In order to see the equation, go into trendline options and click “display equation on chart”. 4. Using this equation determine the concentration of p-Nitrophenol in your Enzyme reaction tubes. Did your enzyme extraction allow the breaking of the β1-4 glycosidic bond between glucose and p-Nitrophenol (p-Nitrophenyl--D glucopyranoside)? Explain. Using excel plot your product concentration (p-Nitrophenol) versus time. Is the plot linear, exponential, does the concentration of product reach a plateau, and not change with any more time? Come up with the chemical reaction for this reaction, using E for the enzyme (cellobiase) and S for the substrate (of p-Nitrophenyl--D glucopyranoside): Thinking in terms of the Enzyme acting as a catalyst, what do you think would happen to your above reaction if you added more enzyme? If you added more substrate? If you heated the reaction? Design an experiment asking one or these questions or one of your own questions, and predict your outcome, if time permits, conduct the experiment. Cellulose is an untapped source of glucose. Glucose is not only extremely important for animal metabolism, but it can be used to make ethanol, which can used in fuel. Currently, glucose from corn is utilized for ethanol production for fuel. Instead of using corn to make biofuel, if we could access the glucose in cellulose, ethanol could be synthesized without impacting animal food sources. Name several sources of cellulose: Explain how you could use cellobiase to generate biofuel. Solutions Extraction buffer and reaction buffer 50 mM MOPS Buffer pH 6 (MW 209.26 g/ mole)-Store at room temp indefinitely Weigh out 2.6 g Dissolve in 150 mL of distilled water Adjust pH to 6 Bring to final volume of 250 mL with distilled water Enzyme Substrate 1.5 mM p-nitrophenyl-B-D glucopyranoside (pNPG) (MW 301.25 g/mole) in 50 mM MOPs Buffer pH 6.0-Store at 4°C, shelf life 1 week. Weigh out 0.023 g Dissolve in 40 mL of 50 mM MOPS Buffer Bring to final volume of 50 mLs with 50 mM MOPS Buffer Stop Solution 200 mM Na2CO3.H2O (MW 124.00 g/mole)-Store at room temp indefinitely Weigh 6.2 g Dissolve in 200 ml of dH2O Bring to final volume of 250 ml with dH2O P-nitrophenol Standard Stock Solution 2.0 mM p-nitrophenol (MW 139.11 g/mole)-Store in at room temp indefinitely Weigh 0.0278 g of p-nitrophenol Dissolve in 75 mL of water Bring to a final volume of 100 mL 0.2 mM p-nitrophenol Standard solution-Store at room temp indefinitely Dilute 2.0 mM p-nitrophenol 1:10 using distilled water by adding 1 mL of 2 mM p-nitrophenol and 9 mL of distilled water in 15 mL conical tube. Dissolve in 75 mL of water Bring to a final volume of 100 mL 0.2 mM p-nitrophenol Standard solution-Store at room temp indefinitely Dilute 2.0 mM p-nitrophenol 1:10 using distilled water by adding 1 mL of 2 mM p-nitrophenol and 9 mL of distilled water in 15 mL conical tube.