sim6364-sup-0001-supplementary
advertisement

Supplementary material for “Accounting for
uncertainty due to ‘Last Observation Carried
Forward’ outcome imputation in a meta-analysis
model”
Vasiliki Dimitrakopoulou, Orestis Efthimiou, Stefan Leucht and
Georgia Salanti
1
Decomposition of the probability of success 𝒑𝒊𝒋 .
Let us denote with P comp ij the probability of a patient randomized in arm j of
study i to complete the study. Also let P impute _ suc ij denote the probability of a
patient to drop out and then be imputed as a success. For the probability pij of a
patient either having an observed or an imputed successful outcome it holds that:
pij P suc comp ij P impute _ suc ij P suc | comp ij P comp ij P impute _ suc ij , (A1)
with:
P impute _ suc ij P impute _ suc true _ suc ij P impute _ suc true _ fail ij
P impute _ suc | true _ suc ij P true _ suc ij
(A2)
P impute _ suc | true _ fail ij P true _ fail ij .
Further, if we denote with P drop ij the probability of a patient to drop out of the
study we have:
P impute _ suc | true _ suc ij
P impute _ suc true _ suc | drop ij P drop ij
P true _ suc ij
,
(A3)
and also:
P impute _ suc | true _ fail ij 1 P impute _ fail | true _ fail ij P comp | true _ fail ij . (A4)
But:
P impute _ fail | true _ fail ij
P impute _ fail true _ fail | drop ij P drop ij
P true _ fail ij
,
(A5)
therefore (A4) becomes:
P impute _ suc | true _ fail ij
1
P impute _ fail
true _ fail | drop ij P drop ij
P true _ fail ij
P comp | true _ fail ij .
We also define sensitivity and specificity as follows:
Seij P impute _ suc | true _ suc, drop ij
P impute _ suc true _ suc | drop ij P drop ij
Spij P impute _ fail | true _ fail , drop ij
P true _ suc | drop ij P drop ij
P impute _ fail true _ fail | drop ij P drop ij
P true _ fail | drop ij P drop ij
and we can write:
P impute _ suc true _ suc | drop ij P drop ij Seij P true _ suc | drop ij P drop ij , (A6)
P impute _ fail
,
true _ fail | drop ij P drop ij Spij P true _ fail | drop ij P drop ij . (A7)
Incorporating Equations (A2) - (A7) into Equation (A1), we obtain:
pij P suc | comp ij P comp ij Seij P true _ suc | drop ij P drop ij P true _ fail ij
Spij P true _ fail | drop ij P drop ij P comp | true _ fail ij P true _ fail ij .
(A8)
Also, we can write the following probabilities:
P true _ suc | drop ij P drop ij P drop | true _ suc ij P true _ suc ij
1 P comp | true _ suc ij P true _ suc ij
P true _ suc ij P comp | true _ suc ij P true _ suc ij
P true _ suc ij P true _ suc | comp ij P comp ij ,
and
P true _ fail | drop ij P drop ij P drop | true _ fail ij P true _ fail ij
1 P comp | true _ fail ij P true _ fail ij
P true _ fail ij P comp | true _ fail ij P true _ fail ij
P true _ fail ij P true _ fail | com ij P comp ij .
,
At this point, we denote the true probability of success as P true _ suc ij ij , and
evidently P true _ fail ij 1 ij . Finally, applying the above into (A8) and after
simplifying, probability p ij is given by:
pij ij Seij Spij 1 P suc | comp ij P comp ij 2 Seij Spij Spij 1 P comp ij 1 .
2
Constraints on Sensitivity and Specificity.
The requirement that 0 ij 1 in Equation (3) of the main paper places constraints
on the values that sensitivity and specificity can take. Also, for Equation
Error! Reference source not found. to be defined the denominator in the right part
of the equation needs be 0 , i.e. Seij Spij 1 0 . This translates into Seij Spij 1
or Seij Spij 1 . We use these constraints to solve for sensitivity and specificity and
we obtain two pairs of inequalities for the permitted values, as follows:
i.
a
For the case that Seij Spij 1 we get Seij max a, b , where:
1
P(suc | com)ij P(com)ij
2 P( suc | com)ij P(com)ij pij P(com)ij 1
Spij 1 P(suc | com)ij P(com)ij P(com)ij
and
b
1
1 P( suc | com)ij P(com)ij
pij 2P(suc | com)ij P(com)ij P(com)ij
.
Sp
P
(
com
)
P
(
suc
|
com
)
1
ij
ij
ij
Also, Spij max c, d , where:
c
1
pij 2 P( suc | com)ij P(com)ij P(com)ij
1 P( suc | com)ij P(com)ij Seij 1 P( suc | com)ij P(com)ij
and
d
ii.
2 P( suc | com)ij P(com)ij pij P(com)ij 1
1
.
P( suc | com)ij 1 P(com)ij 1 Seij P(suc | com)ij P(com)ij
For the case that Seij Spij 1 we get Seij max e, f , where:
e
1
P( suc | com)ij P(com)ij
2 P( suc | com)ij P(com)ij pij P(com)ij 1
Spij 1 P( suc | com)ij P(com)ij P(com)ij
and
f
1
1 P( suc | com)ij P(com)ij
pij 2P(suc | com)ij P(com)ij P(com)ij
.
Sp
P
(
com
)
P
(
suc
|
com
)
1
ij
ij
ij
Also, Spij max g , h , where:
g
1
1 P( suc | com)ij P(com)ij
pij 2 P( suc | com)ij P(com)ij P(com)ij
Seij 1 P( suc | com)ij P(com)ij
and
h
1
2 P( suc | com)ij P(com)ij pij P(com)ij 1
.
Se
P
(
suc
|
com
)
P
(
com
)
P( suc | com)ij 1 P(com)ij 1
ij
ij
ij
3
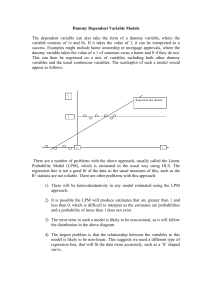
Constructing Figure 1 of the main paper.
Using the results of the analysis presented in the previous Section of this Appendix
we obtain the area of permitted values for sensitivity and specificity for each of the
five available studies. The permitted areas for the studies BENASLP and DRASLP
are presented in Appendix Figure 1 and Appendix Figure 2 respectively. The upper-right
shaded areas correspond to the pairs of permitted values for sensitivity and specificity
for the case that Seij Spij 1 , and the lower left areas to those for the case that
Seij Spij 1 .
Figure 1: Permitted values for sensitivity and specificity (shaded area of the graphs) for
BENASLP as obtained from the relevant inequalities for sensitivity and specificity.
Figure 2: Permitted values for sensitivity and specificity (shaded area of the graphs) for DRASLP
as obtained from the relevant inequalities for sensitivity and specificity.
After producing these areas for all studies, we determine the common area across all
studies. This area corresponds to values of sensitivity-specificity that are permitted for
all studies and it is presented in Figure 1 of the main paper. As already noted in the
main paper, for reasonably stable medical conditions both sensitivity and specificity
are expected to be close to 1 and very low values of Se and Sp are unlikely; therefore
in Figure 1 of the main paper we only focus on the upper area of the graphs that
presents values obtained for the case that Seij Spij 1 .
4 OpenBUGS code
In this section we provide the OpenBUGS code and the data we used for obtaining the
study-specific logORs for the CC analysis, for the analysis of the LOCF-imputed data
and for the meta-analytic model proposed in the main paper. After obtaining the
study-specific logORs for the model of choice, a usual fixed or random effects metaanalysis can be performed to synthesize the findings and obtain the pooled results.
MODELS
### CC ###
model {
for (i in 1:ns){
### binomial likelihood for (observed) successes in completers ###
## arm A
c.s_A[i] ~ dbin(p.suc.com_A[i],c_A[i])
## arm B
c.s_B[i] ~ dbin(p.suc.com_B[i],c_B[i])
p.suc.com_A[i] ~ dbeta(1,1)
p.suc.com_B[i] ~ dbeta(1,1)
theta[i] <- logit(p.suc.com_B[i]) - logit(p.suc.com_A[i])
}
}
### LOCF ###
model {
for (i in 1:ns){
### binomial likelihood for (observed) successes in completers and
### imputed LOCF ###
# arm A
s_A[i] ~ dbin(p_A[i],n_A[i])
# arm B
s_B[i] ~ dbin(p_B[i],n_B[i])
p_A[i]~dbeta(1,1)
p_B[i]~dbeta(1,1)
theta[i] <- logit(p_B[i]) - logit(p_A[i])
}
}
### meta-analytic model with 5 Scenarios of prior settings ###
model {
for (i in 1:ns){
#### binomial likelihood for (observed) successes in imputed LOCF ###
# arm A
succ.drop_A[i] ~ dbin(p.suc.drop_A[i],dropouts_A[i])
p.suc.drop_A[i]~dunif(0,1)
# arm B
succ.drop_B[i] ~ dbin(p.suc.drop_B[i],dropouts_B[i])
p.suc.drop_B[i]~dunif(0,1)
succ_A[i]~dbin(p_A[i],n_A[i])
succ_B[i]~dbin(p_B[i],n_B[i])
p_A[i]~dunif(0,1)
p_B[i]~dunif(0,1)
p.suc.com_A[i]<-(p_A[i]*n_A[i]-dropouts_A[i]*p.suc.drop_A[i])/c_A[i]
p.suc.com_B[i]<-(p_B[i]*n_B[i]-dropouts_B[i]*p.suc.drop_B[i])/c_B[i]
# arm A
c_A[i] ~ dbin(p.comp_A[i],n_A[i])
p.comp_A[i]~dunif(0,1)
# arm B
c_B[i] ~ dbin(p.comp_B[i],n_B[i])
p.comp_B[i]~dunif(0,1)
### decomposed probability of success ###
## arm-specific Se and Sp ##
# arm A
p.tr_A[i] <- max( min( 0.999, ( p_A[i] p.suc.com_A[i]*p.comp_A[i]*(2-Se_A-Sp_A) - (Sp_A-1)*(p.comp_A[i]-1)
)/(Se_A+Sp_A-1)
), 0.001 )
# arm B
p.tr_B[i] <- max( min( 0.999, ( p_B[i] p.suc.com_B[i]*p.comp_B[i]*(2-Se_B-Sp_B) - (Sp_B-1)*(p.comp_B[i]-1)
)/(Se_B+Sp_B-1)
), 0.001 )
theta[i] <- logit(p.tr_B[i]) - logit(p.tr_A[i])
}
### Priors on Sensitivity and Specificity ###
## define the limits of the permitted area ##
Se_low_A <- 0.55 # lower bound for Se of arm A
Se_up_A <- 1
# upper bound for Se of arm A
Se_low_B <- 0.55
Se_up_B <- 1
Sp_low_A <- 0.7
Sp_up_A <- 1
Sp_low_B <- 0.8
Sp_up_B <- 1
# lower bound for Se of arm B
# upper bound for Se of arm B
# lower bound for Sp of arm A
# upper bound for Sp of arm A
# lower bound for Sp of arm B
# upper bound for Sp of arm B
######## chose one of the following Scenarios I-V ########
## Scenario I
## and Sp ##
: Independent Normal priors on the logit scale of Se
# Sensitivity arm A
mean_SE_A <- 2
logit_SE_A ~ dnorm( mean_SE_A, 4)
#generate from a Normal
expit_SE_A <- exp( logit_SE_A ) / ( 1+ exp(logit_SE_A) ) #transform
into normal scale
Se_A <- Se_low_A + (Se_up_A - Se_low_A) * expit_SE_A
#shift to the
area of choice
# Sensitivity arm B
mean_SE_B <- 2
logit_SE_B ~ dnorm( mean_SE_B, 4 )
#generate from a Normal
expit_SE_B <- exp( logit_SE_B ) / ( 1+ exp(logit_SE_B) ) #transfrom
into normal scale
Se_B <- Se_low_B + (Se_up_B - Se_low_B) * expit_SE_B
#shift to the
area of choice
# Specificity arm A
mean_SP_A <- 2
logit_SP_A ~ dnorm( mean_SP_A, 4 )
#generate from a Normal
expit_SP_A <- exp( logit_SP_A ) / ( 1+ exp(logit_SP_A) )
#transform
into normal scale
Sp_A <- Sp_low_A + (Sp_up_A - Sp_low_A) * expit_SP_A
#shift to the
area of choice
# Specificity arm B
mean_SP_B <- 2
logit_SP_B ~ dnorm( mean_SP_B, 4 )
#generate from a Normal
expit_SP_B <- exp( logit_SP_B ) / ( 1+ exp(logit_SP_B) )
#transform
into normal scale
Sp_B <- Sp_low_B + (Sp_up_B - Sp_low_B) * expit_SP_B
#shift to the
area of choice
## Scenario II : Independent Uniform priors ##
Se_A ~dunif(0.55,1)
Sp_A ~dunif(0.7,1)
Se_B ~dunif(0.55,1)
Sp_B ~dunif(0.8,1)
## Scenario III : Independent Beta priors ##
rand_se_A ~ dbeta(2,2)
rand_sp_A ~ dbeta(2,2)
rand_se_B ~ dbeta(2,2)
rand_sp_B ~ dbeta(2,2)
Se_A <- Se_low_A + (Se_up_A - Se_low_A) * rand_se_A
#Sensitivity arm A
Sp_A <- Sp_low_A + (Sp_up_A - Sp_low_A) * rand_sp_A
#Specificity arm A
Se_B <- Se_low_B + (Se_up_B - Se_low_B) * rand_se_B
#Sensitivity arm B
Sp_B <- Sp_low_B + (Sp_up_B - Sp_low_B) * rand_sp_B
#Specificity arm B
## Scenario IV : Independent truncated Beta priors ##
Se_A ~ dbeta(2,2) T(0.55,)
Sp_A ~ dbeta(2,2) T(0.7,)
Se_B ~ dbeta(2,2) T(0.55,)
Sp_B ~ dbeta(2,2) T(0.8,)
## Scenario V : Independent Normal priors on the logit scale of Se
and Sp ##
#Sensitivity arm A
mean_SE_A <- -2
logit_SE_A ~ dnorm( mean_SE_A ,4 )
#generate from a Normal
expit_SE_A <- exp( logit_SE_A ) / ( 1+ exp(logit_SE_A) )
#transform into normal scale
Se_A <- Se_low_A + (Se_up_A - Se_low_A) * expit_SE_A
#shift to the
area of choice
# Sensitivity arm B
mean_SE_B <- -2
logit_SE_B ~ dnorm( mean_SE_B, 4 ) #generate from a Normal
expit_SE_B <- exp( logit_SE_B ) / ( 1+ exp(logit_SE_B) )
#transform
into normal scale
Se_B <- Se_low_B + (Se_up_B - Se_low_B) * expit_SE_B
#shift to the
area of choice
# Specificity arm A
mean_SP_A <- -2
logit_SP_A ~ dnorm( mean_SP_A, 4 ) #generate from a Normal
expit_SP_A <- exp( logit_SP_A ) / ( 1+ exp(logit_SP_A) )
#transform
into normal scale
Sp_A <- Sp_low_A + (Sp_up_A - Sp_low_A) * expit_SP_A
#shift to the
area of choice
# Specificity arm B
mean_SP_B <- -2
logit_SP_B ~ dnorm( mean_SP_B, 4 )
#generate from a Normal
expit_SP_B <- exp( logit_SP_B ) / ( 1+ exp(logit_SP_B) )
#transform
into normal scale
Sp_B <- Sp_low_B + (Sp_up_B - Sp_low_B) * expit_SP_B
#shift to the
area of choice
}
DATA
list(
## number of studies ##
ns = 5,
## randomised patients in arms A and B ##
n_A=c( 71, 255, 95, 368, 97),
n_B=c( 62, 64, 96, 118, 105),
dropouts_A=c(19,60,24,33,6),
dropouts_B=c(24,21,37,10,15),
## completers in arms A and B ##
c_A=c( 52, 195, 71, 335, 91),
c_B = c( 38, 43, 59, 108, 90),
## successes in completers in arms A and B ##
c.s_A = c( 41, 146, 51, 142, 55),
c.s_B = c( 29, 36, 34, 33, 41),
succ.drop_A=c(8,6,3,2,0),
succ.drop_B=c(7,0,5,0,1),
succ_A=c(49, 152, 54, 144, 55),
succ_B=c( 36, 36, 39, 33, 42)
)