mec13347-sup-0001-FigS1-S4-TableS1-S2
advertisement

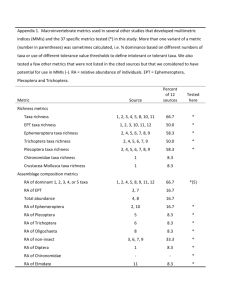
Supplementary information Table S1. Statistics of linear models describing the relationships between the logarithmic transformation of bacterial production (leucine uptake in pmol L-1 h-1) and community diversity estimates. Estimates for the active fraction and standing stock are based on the 16S rRNA and 16S rRNA gene respectively. H: Shannon index, SES: standardized phylogenetic diversity. Significant relationships are in boldface. Active fraction Standing stock Variable Intercept Slope df F R2 P value H Richness SES 1.23 1.32 1.12 -0.54 -0.00 0.21 39 39 39 0.60 2.57 10.86 0.02 0.06 0.22 0.44 0.12 <0.01 H Richness SES 1.8 1.68 1.04 -0.37 -0.01 0.03 44 44 44 14.01 21.28 0.15 0.24 0.33 0.01 <0.01 <0.01 0.70 Table S2. Permutation test under reduced model for canonical correspondence analysis (CCA) of community assemblies against environmental conditions (salinity, chlorophyll a, nitrate, nitrite, ammonium, phosphate and silicate) for the active fraction of the community and the standing stock. Chisq df F R2 P value Active fraction 1.67 8 1.25 0.03 0.001 Standing stock 1.43 8 1.16 0.00 0.021 1.0 Bacterial production 0.0 0.5 1.0 0.5 0.0 Bacterial production 1.5 b 1.5 a 50 75 100 30 Community richness a 60 90 Community richness b Figure S1. Relationship between bacterial production and the community richness for the active bacteria (a) and the standing stock (b). Bacterial production is measured as the rate of tritiated-leucine incorporation (pmol L-1 h-1) and plotted following logarithmic transformation. The black line in panel (b) represents a linear fit to the data (R2=0.33, P<0.01, n=46) and the grey shading shows the 95% confidence interval. Figure S2. Relationship between bacterial production and the phylogenetic diversity of active bacteria without weighting by taxa abundance. Without weighting by taxa, all the rare bacteria become as important as the abundant ones in the calculations. In theory, if rare taxa can be find everywhere, the phylogenetic diversity of communities will not vary much when taxa abundance is not taken into account. We thus argue that in the case of natural marine bacteria communities, characterized by few abundant taxa and a very large number of rare taxa, phylogenetic diversity has to be weighted by taxa abundance. a b Figure S3. Differences in bacterial production in relation to similarity between communities. Differences in bacterial production are calculated as Euclidian distance between each pair of samples and community similarity as Bray-Curtis (a) or Sorensen (b) index of similarity. The grey shading shows the 95% confidence interval. Figure S4. Periodogram obtained by spectral analysis showing the periodicity of community diversity for the standing stock (DNA) and the active bacteria fraction (RNA). Horizontal lines show the white noise lines illustrating significance level of 0.01 and 0.05. DNA shows a significant pattern of diversity with a frequency of 1 year while RNA has no significant pattern.