Some detailed procedures - Springer Static Content Server
advertisement

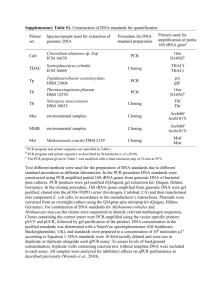
ELECTRONIC SUPPLEMENARY MATERIAL Some detailed procedures We used 0.25 and 1.0 g soil for DNA and RNA extraction, respectively. The extraction was conducted using the PowerSoil™ DNA Isolation Kit and PowerSoil® Total RNA Isolation Kit (MO BIO Laboratories, Carlsbad, CA, USA), as described by suppliers. After the removal of DNA residuals using DNase I (MO BIO Laboratories, Carlsbad, CA, USA), RNA extracts were reverse-transcribed into cDNA using the PrimeScript™II 1st Strand cDNA Synthesis Kit with random hexamers (Takara Bio Inc., Shiga, Japan). Then, DNA and cDNA solutions were diluted 5 and 50 times, respectively, for subsequent operations. Copies of 16S rDNA and its transcripts were determined using an 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) with universal bacterial 16S rRNA gene primers 338F (ACT CCT ACG GGA GGC AG, Lane et al. 1991), and 518R (ATT ACC GCG GCT GCT GG, Muyzer et al. 1993). The 20 μl qPCR reaction system contained 1 μl template DNA (DNA, cDNA, or 10-fold serially diluted standard), 10 μl Maxima™ SYBR Green/ROX (2 ×, Fermentas, Waltham, MA, USA), 0.5 μl forward primer (20 μmol l-1), 0.5 μl reverse primer (20 μmol.l-1), 0.25 μl bovine serum albumin (25 mg ml−1; Promega, Madison, WI, USA), and 7.75 μl nuclease-free water. Standard curves were constructed using plasmids harboring the 16S rRNA gene fragment. All DNA, cDNA, and standards were analysed in triplicate, and three no template controls were used to check the contaminations of reagents. PCR runs started with an initial denaturation and enzyme activation step for 10 minutes at 95 °C, followed by 40 cycles of 15 s at 95 °C, 30 s at 56 °C, 30 s at 72 °C, and 15 s at 80 °C. We recorded the fluorescence signal at 80 °C to attenuate the influence of primer dimers. The specificity of PCR products was verified by melting curve analysis. The qPCR efficiencies were around 95 %, and the Standard curve R2 > 0.999. The absence of PCR inhibitors in DNA and cDNA solutions was checked via qPCR with dilution series of corresponding solutions (Hargreaves et al. 2013). Terminal restriction fragment polymorphism (T-RFLP) analysis was conducted with universal bacterial 16S rRNA gene primers 27F (AGA GTT TGA TCM TGG CTC AG, Giovannoni et al. 1991) and 519R (GWA TTA CCG CCG CKG CTG, Lane et al. 1985). The forward primer was labelled with the fluorescent dye FAM (6carboxyfluorescein) at the 5’ end. The 50 μl PCR reaction system contained 2.5 μl template DNA (or cDNA) solution, 25 μl Premix Taq™ Hot Start Version (Takara Bio Inc., Shiga, Japan), 1 μl forward primer (20 μmol l- 1 1 ), 1 μl reverse primer (20 μmol l-1), 0.5 μl bovine serum albumin (25 mg ml−1; Promega, Madison, WI, USA), and 20 μl nuclease-free water. PCR programs were as follows: initial denaturation and enzyme activation at 95 °C for 10 minutes, followed by 30 cycles of 45 s at 95 °C, 40 s at 56 °C, 55 s at 72 °C, and ending with a final extension at 72 °C for 10 minutes. All the samples were run in duplicate and the duplicate PCR products were pooled for the following operations. After being purified using the Agarose Gel DNA Purification Kit (Axgen Biotechnology Corporation, Hangzhou, China), the purified PCR products were completely digested by restriction endonuclease ALU Ⅰ (New England Biolabs Inc. Beverly, MA, USA). Terminal restriction fragments were size-separated by the Beijing Tsingke BioTeck Co. Ltd (Beijing, China) using the 3730xl DNA Analyser (Applied Biosystems®, Foster City, CA, USA). Then, the electropherograms were retrieved using Peak Scanner 1.0 (Applied Biosystems®, Foster City, CA, USA) and the threshold for peak assignment was +/0.5 bp. After uniformization, unqualified peaks with size < 30 bp, > 540 bp or peak height < 20 were removed. Proportions of peaks were calculated according their peak areas. Then, the proportions were arcsine transformed for the following data analysis. References Giovannoni SJ (1991) The polymerase chain reaction. In: Stackebrandt E, Goodfellow MD (ed) Nucleic acid techniques in bacterial systematics, 1st edn. Wiley, New York, pp 177-203 Hargreaves SK, Roberto AA, Hofmockel KS (2013) Reaction- and sample-specific inhibition affect standardization of qPCR assays of soil bacterial communities. Soil Biol Biochem 59:89-97 Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow MD (ed) Nucleic acid techniques in bacterial systematics,1st edn. Wiley, New York, pp 115-175 Lane DJ, Pace B, Olsen GJ, Stahl DA, Sogin ML, Pace NR (1985) Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci 82:6955-6959 Muyzer G, Dewaal EC, Uitterlinden AG (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59:695-700 2