jbi12527-sup-0001-AppendixS1-S3
advertisement
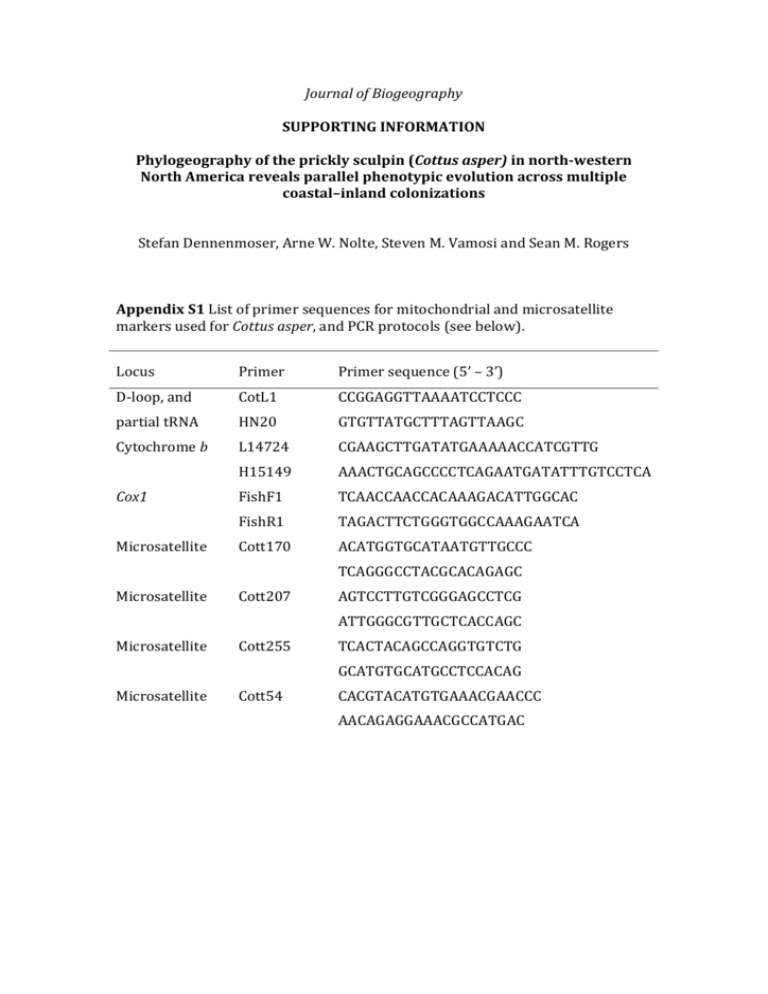
Journal of Biogeography SUPPORTING INFORMATION Phylogeography of the prickly sculpin (Cottus asper) in north-western North America reveals parallel phenotypic evolution across multiple coastal–inland colonizations Stefan Dennenmoser, Arne W. Nolte, Steven M. Vamosi and Sean M. Rogers Appendix S1 List of primer sequences for mitochondrial and microsatellite markers used for Cottus asper, and PCR protocols (see below). Locus Primer Primer sequence (5’ – 3’) D-loop, and CotL1 CCGGAGGTTAAAATCCTCCC partial tRNA HN20 GTGTTATGCTTTAGTTAAGC Cytochrome b L14724 CGAAGCTTGATATGAAAAACCATCGTTG H15149 AAACTGCAGCCCCTCAGAATGATATTTGTCCTCA FishF1 TCAACCAACCACAAAGACATTGGCAC FishR1 TAGACTTCTGGGTGGCCAAAGAATCA Cott170 ACATGGTGCATAATGTTGCCC Cox1 Microsatellite TCAGGGCCTACGCACAGAGC Microsatellite Cott207 AGTCCTTGTCGGGAGCCTCG ATTGGGCGTTGCTCACCAGC Microsatellite Cott255 TCACTACAGCCAGGTGTCTG GCATGTGCATGCCTCCACAG Microsatellite Cott54 CACGTACATGTGAAACGAACCC AACAGAGGAAACGCCATGAC Microsatellite CottE30 AAGCGGACAGACGCAAAGAG ACTCGCCCAAGAAGACCAGC Microsatellite LCE54 ATGTGTGAGGGCCTCAATGC CTGTGTGGTGCGTCCATTGG Microsatellite Cott214 AGTAGTGCCGCTGCACCCAC TCTCCCTCGTGGTGCAAACC Microsatellite Cott78 TCTGCTCCGCAGGGTCGCTG TGACACATGGTGAACCCGTG Microsatellite Cott348 TGGACCACTGAGTTGCTGTG TGGTGTGTCATACCTGCAGC Microsatellite CottE11 TCAAGAGGTGAGAGGCCAAGAG CATCCTGGTGTTACCATTGCTG Microsatellite CottE20 CGTCTGCACAGTTGAGCCTG AGCACGCCGTCGCTAACAGC Microsatellite Cott153 GGGTTCAGGTGCGTTTGGTC ACCTTCAGCTCCGACATCAG Microsatellite Cott213 TTGCCATGGATTTGAGGCAG AGCATTGCTATTATCAGGCTGC Microsatellite CottE13 AGTTACGCAATGCAACACGCCT TCGAACACGAAGCTCCCGTCAC Microsatellite LCE81 GCTTGTGAACATTTCGGCCTC AATGTACAAAGCTCCCACGTCC Microsatellite Cott50 TCCTGTGCTAATGCTGTGGA AAACCTCACCTCGGTGAGTC Microsatellite Cott686 TGGGAGTGCGATCGGGTTGG GCGGGCCTTGACACTGTTGG Microsatellite CottE23 TGAGCAGCTTTACAGTGTTGG TCGTGCTCTTCTGGCATCTG Microsatellite EDA1 GGACAGCACACTCCTTCCAT AGAGGCCCTTAAAAGGCAGA PCR protocols used in this study: For obtaining mitochondrial DNA sequences, we applied the following primer combinations: CotL1 and HN20 (partial t-RNA, D-loop, Šlechtová et al., 2004), L14724 (Meyer et al., 1990) and H15149 (Kocher et al., 1989, cytochrome b), FishF1 and FishR1 (Cox1; Ward et al., 2005). Amplification of D-loop sequences (CotL1/HN20, 583 bp) was performed in 30-L volumes containing the following final reaction concentrations: 1× PCR-Buffer, 2mM MgCl2, 0.2 mM of each dNTP, 10 pmol of each primer, 0.625 units of TAQ-polymerase, and ~200 ng genomic DNA. The PCR protocol consisted of 2 minutes at 95C, followed by five cycles of [94C -45s, 48C -45s, 72C -90s], 29 cycles of [94C -45s, 52C -45s, 72C -90s], and 5 minutes at 72C final elongation. For Cox1 sequences (FishF1/R1, 583 bp), PCR reactions were performed in 25-L volumes containing 1× PCR-Buffer, 2mM MgCl2, 0.2 mM of each dNTP, 10 pmol of each primer, 0.625 units of TAQ-polymerase, and ~200 ng genomic DNA. Cycling conditions consisted of 95C for 2 minutes, followed by 35 cycles of [94C -30s, 52C -40s, 72C -60s], and a final elongation of 72C for 10 minutes. A cytochrome b fragment (419 bp) was amplified with the L14724/H15149 primer pair in 30-l volumes containing 1× PCR-Buffer, 2mM MgCl2, 0.2 mM of each dNTP, 10 pmol of each primer, 0.625 units of Taq-polymerase, and c. 200 ng genomic DNA. Cycling conditions consisted of 95C for 2 minutes, followed by 35 cycles of [95C -50s, 50C -15s, 72C -80s], and a final elongation of 72C for 7 minutes. Microsatellites were multiplexed with up to six fluorescently-labelled primers using QIAGEN Multiplex Kits (Qiagen, Inc.), and amplified in 10 L-reactions (5 L Qiagen-Multiplex-Kit, 0.3-0.6 M primer, 40 ng DNA-template). PCR-protocols consisted of an initial denaturation step of 95C for 15 minutes, followed by 38 cycles of [94C -30s, 60C -90s, 72C -60s], and a final elongation of 72C for 30 minutes. REFERENCES Kocher, T.D., Thomas, W.K., Meyer, A., Edwards, S.V., Pääbo, S., Villablanca, F.X. & Wilson, A.C. (1998) Dynamics of mitochondrial DNA evolution in animals: amplification and sequencing with conserved primers. Proceedings of the National Academy of Sciences USA, 86, 6196-6200. Meyer, A., Kocher, T.D., Basasibwaki, P. & Wilson, A.C. (1990) Monophyletic origin of Lake Victoria cichlid fishes suggested by mitochondrial DNA sequences. Nature, 347, 550-553. Šlechtová, V., Bohlen, J., Freyhof, J., Persat, H. & Delmastro, G.B. (2004) The Alps as barrier to dispersal in cold-adapted freshwater fishes? Phylogeographic history and taxonomic state of the bullhead in the Adriatic freshwater drainage. Molecular Phylogenetics and Evolution, 33, 225–239. Ward, R.D., Zemlak, T.S., Innes, B.H., Last, P.R. & Hebert, P.D.N. (2005) DNA barcoding Australia’s fish species. Philosophical Transactions of the Royal Society Series B: Biological Sciences, 360, 1847–1857. Appendix S2 Prickling and body shape differences between inland and coastal sculpins. Figure S2 A) Landmark configuration used for Cottus asper (1 = tip of nasal, 2 = interorbital pore, 3 = end of head, 4 = first dorsal fin origin, 5 = second dorsal fin origin, 6 = end of second dorsal fin, 7 = upper caudal fin origin, 8 = midpoint caudal, 9 = lower caudal fin origin, 10 = anal fin end, 11 = anal fin origin, 12 = lower pectoral fin origin, 13 = upper pectoral fin origin, 14 = upper origin of gill opening, 15 = opercular spine, 16 = 3rd pore of the preopercular-mandibular canal, 17 = posterior end of maxilla, 18 = posterior end of eye, 19 = midpoint of eye, 20 = distal end of eye, 21 = nasal nostril, 22 = midpoint between 7 and 9, 23 = mid of caudal peduncle). B) Wireframe graph showing shape differences between coastal (black line) and inland (grey line) groups (scale factor × 6). C) Frequencies (circle sizes represent percentages) of prickling categories (high, medium, low) in coastal and inland populations. D) PCA plot visualizing shape variation among coastal (black) and inland (grey) individuals along the first two principal components (with 95% confidence ellipses). Appendix S3 Fifty per cent majority-rule consensus tree of mtDNA sequences. Figure S3 Fifty per cent majority-rule consensus tree of mtDNA sequences of Cottus asper. Numbers above branches indicate Bayesian inference posterior probabilities, and numbers below branches maximum likelihood bootstrap proportions. Genetic lineages inferred from tree clusters are indicated by vertical bars (black = A, grey = B, white = C).









