Development and characterization of novel microsatellite loci for
advertisement

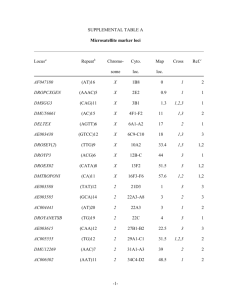
Development and characterization of novel microsatellite loci for Pinus yunnanensis from transcriptome using next-generation sequencing Nianhui Cai1, Yulan Xu1,2, Yang Xu1,2, Dawei Wang1,2, Bin Tian2, Chengzhong He1,2, Anan Duan1 1 Southwest Forestry University, Key Laboratory for Forest Genetic and Tree Improvement & Propagation in Universities of Yunnan Province, Kunming 650224, Yunnan, China; E-Mails: cainianhui@sohu.com (N. C.) 2 Key Laboratory of Biodiversity Conservation in Southwest China, State Forestry Administration, Southwest Forestry University, Kunming, 650204, China E-Mails: xvyulan@163.com(Y. X.); 1160967757@qq.com; warpunk@163.com (D. W.); tianbinlzu@gmail.com (B. T.); hcz70@163.com(C. H.); duananan@gmail.com(A. D.) Abstract Pinus yunnanensis Franch., is a major component of coniferous forests in southwestern China, has experienced the excellent resources declines. Nevertheless, little is known about its intraspecific genetic variation. We identified a set of 32 novel microsatellite loci for P. yunnanensis though nextgeneration sequencing and tested amplification, of which 24 were amplified successfully for the species. Markers were screened for the assessment of polymorphism in 24 individuals sampled from four natural populations of P. yunnanensis and 21 of them were polymorphic. The number of alleles per locus varied from 1 to 8 with a mean of 3.3. The observed and expected heterozygosity ranged from 0.000 to 1.000 and 0.000 to 0.861, with averages of 0.506 and 0.495, respectively. No significant linkage disequilibrium was detected in any pair-wise loci. These novel polymorphic microsatellite loci would be useful for population genetic studies of P. yunnanensis to inform the conservation and management strategies. Keywords: Pinus yunnanensis; Microsatellite; Population genetics; Next generation sequencing Pinus yunnanensis French. is extensively used in reforestation of great economic and ecological importance in Southwest China. It grows at altitudes of 600 to 3100 m and ranges from 23° to 30° N and 96° to 108° E (Wu 1986). Unfortunately, human overexploitation and natural disasters such as fire and drought have seriously damaged the P. yunnanensis resource. The knowledge of the level and distribution of intra-specific variation is necessary to better understand population genetics and inform the conservation activities and sustainable use of genetic resources (Liu and Hammett 2014). There are few descriptions of the intra-specific variability of P. yunnanensis, but they have shown that morphological traits varied significantly among populations or provenances. Little is known about the extent and pattern of intra-specific variation of P. yunnanensis at the molecular level. The aim of the present study was to identify novel microsatellite markers by analyzing a transcriptome de novo assembled from RNA-seq data for P. yunnanensis to adequately assess genetic variation and make conservation strategies. Microsatellites for P. yunnanensis were identified from a next-generation sequencing run. Eighty thousands Unigene were analyzed using MicroSAtellite (MISA) to find reads contained di-, tri, tetra-, penta- and hexa-nucleotide repeats and 2899 SSR primers were designed using Primer Premier 5.0 program with default settings. 32 potential microsatellites were randomly chosed to test amplification using three individuals. Of them 24 microsatellite markers were amplified successfully and then were screened for the assessment of polymorphism in 24 individuals from four populations located different regions across main distribution areas. Total genomic DNA was extracted from fresh needles following a modified cetyl trimethyl ammonium bromide (CTAB) protocol (Doyle and Doyle, 1990). The PCR reaction volume was 30 μl using a Takara Taq mixture kit (Takara, Dalian, Liaoning, China), PCR products were separated by capillary electrophoresis using an ABI3730xl DNA Analyzer (Applied Biosystems). The data were performed using the software GeneMarker V2.2.0 with the LIZ500 size standard. The number of alleles (Na), and observed (Ho), expected heterozygosity (He), Hardy-Weinberg equilibrium in each loci and pair-wise loci linkage disequilibria (LD) were evaluated for each population using POPGENE v 1.32 (Yeh et al. 1997). Of them, 21 microsatellites showed polymorphism (Supplementary Material: Table S1). The number of alleles per locus ranged from 1 to 8 with an average of 3.3. The observed and expected heterozygosity varied from 0.000 to 1.000, and from 0.000 to 0.861 with means of 0.506 and 0.495, respectively (Supplementary Material: Table S2). PyTr27 loci showed significant deviation from Hardy-Weinberg equilibrium. No locus pair revealed significant linkage disequilibrium in all of populations, implying that all loci were not closely linked to each other, thus making them suitable for assessing the genetic variation of this species. These 21 novel polymorphic loci will add the relatively small number of eight existing microsatellites described previously in this species (Xu et al. 2013). Acknowledgements This study was supported by grants from the National Natural Science Foundation of China (NSFC No. 31360189 and 31260191). References Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12(1):13-15. Liu JJ, Hammett C (2014) Development of novel polymorphic microsatellite markers by technology of next generation sequencing in western white pine. Conservation Genetics Resources 6(3): 647-648. Wu ZY (1986)Flora Yunnanica (Tomus 4): Spermatophyta. Science Press, Beijing, pp 54-57. Xu YL, Zhang RL, Tian B, Bai QS, Wang DW, Cai NH, He CZ, Kang XY, Duan AA (2013) Development of novel microsatellite markers for Pinus yunnanensis and their cross amplification in congeneric species. Conservation Genetics Resources 5(4): 1113-1114. Yeh FC, Yang RC, Boyle T (1997) PopGene Version 1.31, Microsoft Window-Based Freeware for Population Genetics Analysis: Quick User Guide. University Alberta/Center for International Forestry Research, Canada, pp 1-28.