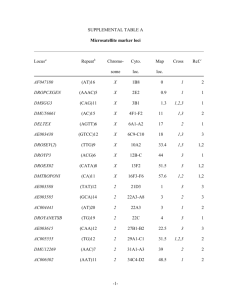
Appendix A2 Genetic structure of E. globulus samples The genetic
advertisement

Appendix A2 Genetic structure of E. globulus samples The genetic structure among the sample of E. globulus collected from the discovery population was investigated using the computer program STRUCTURE and a data set derived from 18 microsatellite loci. Of these microsatellites, six (CRC2, CRC5, CRC7, CRC8, CRC10 and CRC 11) were designed by Steane et al. (2001), two (EMBRA 10 and EMBRA 11) by Brondani et al. (1998), four (EMBRA23, EMBRA30, EMBRA38 and EMBRA63) by Brondani et al. (2002), two (EMBRA 210 and EMBRA 213) by Brondani et al. (2006), one (Eg131) by Thamarus et al. (2002) and three (EMBRA362, EMBRA712 and EMBRA747) by D. Grattapaglia (pers. comm.). Multiplex PCRs were carried out using a PCR cycle comprising: 15 minutes at 95°C, 30 cycles of 30 seconds at 94°C, 90 seconds at 57°C, 1 minute at 72°C; followed by a final 10 minutes at 60°C. The PCR products were separated on an ABI 377 DNA sequencer at the Australian Genomic Research Facility (AGRF) in Adelaide and analysed using GeneMapper® software (Version 3.7). The SSR loci were highly polymorphic and informative with on average 21.7 alleles per loci. Figure 1. Graph of ΔK calculated from 18 microsatellites for E. globulus microsatellite following the method of Evanno et al. (2005). Evanno et al. (2005) provides a method for finding the most likely number of clusters of genetically homogeneous groups of individuals (K). The secondary peak at K=4 was deemed the best solution as it made the most biological sense. Figure 2 Proportion of membership at K=4 for all sampled individuals of E. globulus from the discovery population based on microsatellite data. Each individual is represented by a thin vertical line that can be partitioned into up to four coloured segments representing the individual’s estimated membership (Q) into the K genetic clusters. Individuals are grouped by region (above the figure) and race (below the figure). References Brondani R, Williams E, Brondani C, Grattapaglia D (2006) A microsatellite-based consensus linkage map for species of Eucalyptus and a novel set of 230 microsatellite markers for the genus. Bmc Plant Biology 6 (1):20 Brondani RPV, Brondani C, Grattapaglia D (2002) Towards a genus-wide reference linkage map for Eucalyptus based exclusively on highly informative microsatellite markers. Molecular Genetics and Genomics 267:338-347 Brondani RPV, Brondani C, Tarchini R, Grattapaglia D (1998) Development, characterization and mapping of microsatellite markers in Eucalyptus grandis and E. urophylla. Theoretical and Applied Genetics 97 (5-6):816-827 Steane DA, Vaillancourt RE, Russell J, Powell W, Marshall D, Potts BM (2001) Development and characterisation of microsatellite loci in Eucalyptus globulus (Myrtaceae). Silvae Genetica 50 (2):89-91 Thamarus KA, Groom K, Murrell J, Byrne M, Moran GF (2002) A genetic linkage map for Eucalyptus globulus with candidate loci for wood, fibre, and floral traits. Theoretical and Applied Genetics 104 (2-3):379-387