Maize Single Site Analysis
advertisement
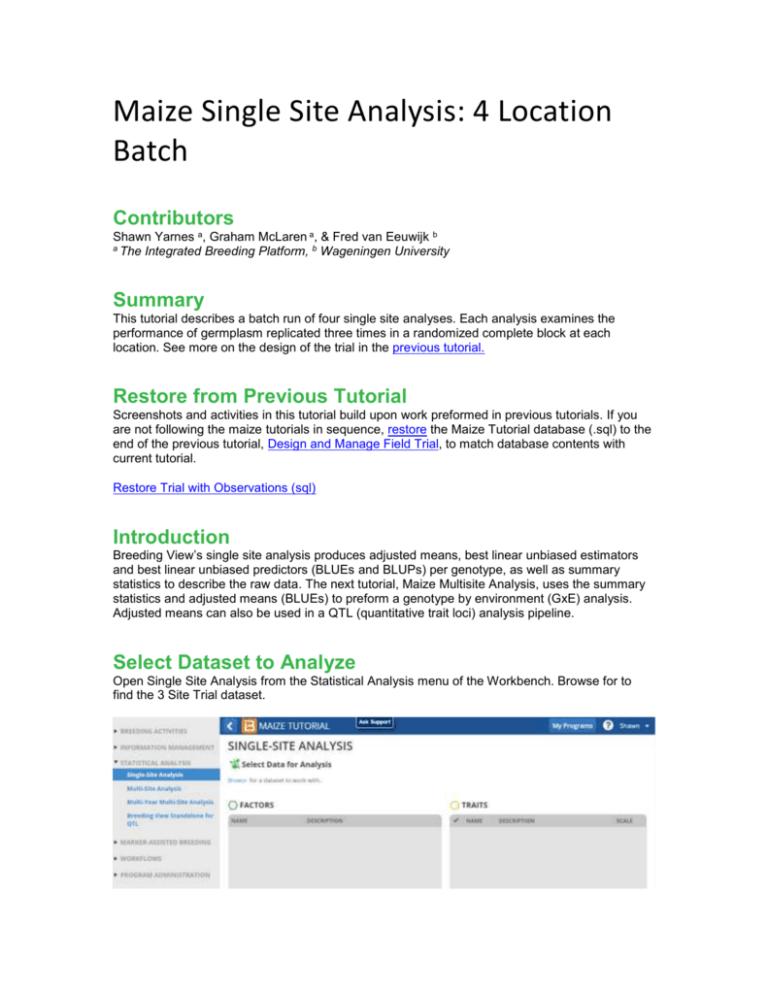
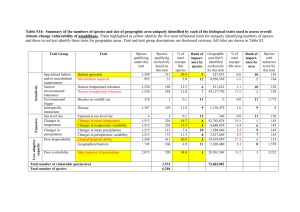
Maize Single Site Analysis: 4 Location Batch Contributors Shawn Yarnes a, Graham McLaren a, & Fred van Eeuwijk b a The Integrated Breeding Platform, b Wageningen University Summary This tutorial describes a batch run of four single site analyses. Each analysis examines the performance of germplasm replicated three times in a randomized complete block at each location. See more on the design of the trial in the previous tutorial. Restore from Previous Tutorial Screenshots and activities in this tutorial build upon work preformed in previous tutorials. If you are not following the maize tutorials in sequence, restore the Maize Tutorial database (.sql) to the end of the previous tutorial, Design and Manage Field Trial, to match database contents with current tutorial. Restore Trial with Observations (sql) Introduction Breeding View’s single site analysis produces adjusted means, best linear unbiased estimators and best linear unbiased predictors (BLUEs and BLUPs) per genotype, as well as summary statistics to describe the raw data. The next tutorial, Maize Multisite Analysis, uses the summary statistics and adjusted means (BLUEs) to preform a genotype by environment (GxE) analysis. Adjusted means can also be used in a QTL (quantitative trait loci) analysis pipeline. Select Dataset to Analyze Open Single Site Analysis from the Statistical Analysis menu of the Workbench. Browse for to find the 3 Site Trial dataset. Select the CIMMYT 2012 Trial. Review the factors and make sure the 7 phenotypic traits measured in this trial dataset are selected, and click Next. Specify Analysis Conditions Use the default analysis name. Select Incomplete Block Design for the design type. The factor that defines environment is LOCATION_NAME. Select all four locations to preform individual single site analyses on each location. REP_NO is the factor in this dataset that defines replications. DESIGNATION defines the germplasm factor to be used in the analysis. Click Run Breeding View to launch the breeding view application. Run Analysis When the Breeding View application launches the analysis conditions and data are loaded. Notice that the four locations and all seven traits are selected with green checks. The analysis pipeline includes a set of connected nodes, which can be used to run and configure pipelines. Leave the default settings for the pipeline and run. Right click the first box and Run Selected Environment Pipelines. When the analysis is complete a popup notifies the user. Quality Assurance Breeding View provides an overview of potentially influential measurements to help users identify and possibly exclude observations. Many influential observations will reflect true genotypic variation and these data should not be excluded from the trial. The only observations that deserve exclusion are obvious errors or measurements influenced by heterogenous environmental variation within a block, like damage to a single plot. Select and open the Sabana Del Medio report. The table displays potential influential observations identified by the raw data method, which identifies observations exceeding 1.5 times the interquartile range, and the residual method, which identified standardized residuals by mixed model analysis. Select the potential outlier, plant height (PH) with the value of 95. Review the box plot for this trait as well as the related measurements for this genotype. Exclude Observation The measurement of 95 is well below the height measurements for other replicates of this genotype, indicating a possible error in the phenotypic observation. Exclude this anomalous value from the analysis by selecting Set Selected to Missing. Re-Run Analysis Return to the analysis pipeline and run again. Notice the boxplot for PH at Sabana Del Medio has changed in the Quality Assurance report. The adjusted means and heritability values for this trait will have also changed in the trial Report. Select Upload to the BMS to save the adjusted means and summary statistics to the BMS database for use in a subsequent genotype by environment (GxE) analysis. Analysis Report This section of the tutorial provides a brief guide to interpretation of the Report tab, which includes analysis results, including graphs. The report tab summarizes the individual single analyses preformed in batch. The majority of the results are included within the individual trial reports. Report Tab Contents Heritability Table Combined file of predicted means: Excel file of BLUEs and BLUPs Links to individual trial reports Heritability Table The values presented in the heritability table summarize the heritabilities calculated for each trait by location. Broad-sense mean line heritability is derived from an estimate of the correlation between the genotype BLUPs and their unknown true value (Cullis, Smith & Coombes, 2006). If a model cannot be fitted to the trait data, such when there is no variability in the trait measurement, that trait will be missing from the heritability table. Heritability Values: Notice that some of the seven traits are missing from some or all of the locations. For example, average shelling percentage (aSP) is missing from all locations. There was no variation measured in this trait at any location. No adjusted means could be calculated for this trait, because all entries were recorded as 0.788, making this trait non-informative. Combined File of Predicted Means Select the link to the combined file of predicted means and open in Excel. Non-informative traits that could not be fitted with a model will appear as missing data. BLUPs across all locations: Notice that there is no column for average shelling percentage (aSP), because models could not be fitted to the aSP data at any location. Two traits, grain moisture (MOI) and anthesis date (AD), at San Gilberto have empty cells for the same reason. Individual Trial Reports Although the four single site analyses are summarized together in the heritability table, each location is represented by an individual analysis. Select the link to the Tlaltizapan individual trial report to review the analysis performed at this location. Individual trial reports include: File of predicted means: Link to Excel (.xls) file containing an environment-specific subset of BLUEs and BLUPs presented in the combined file of predicted means. 20 Best Genotypes Table: This table is 20 best genotypes as defined by BLUPs sorted by the first trait. If you are interested in sorting genotypes by other traits, you can scroll down to view the analysis of individual traits or perform a custom sort of BLUPs within the file of predicted means (.xls). Summary of Traits: A table presenting the minimum, mean, maximum, and heritability for each trait within this location based on BLUPs Estimated Genetic Correlations between traits Principle Components Biplot Individual Trait Analyses: Summary statistics of raw data, heritability, sorted genotype table, summary statistics based on BLUPs, phenotypic correlations, principle components biplot File of Predicted Means Tlaltizapan Report Summary: The beginning of each report provides the project name, the environment name, the field design, and the date. Users are presented a link to the adjusted means data for this environment, which is a subset of the data presented in the combined mean file reporting all locations. Users are also notified about analysis failures. In this case average shelling percentage (aSP) failed to be fitted with a mixed model, because this trait had no reported variation within this location. Best Genotypes Sorted by BLUPs Tlaltizapan 20 Best Genotypes Sorted by AD: This table is the 20 best genotypes sorted by BLUPs for the first trait, anthesis date, as defined by the default Analysis Pipeline settings under the Generate Report node. The default settings sort values in ascending sort with large values interpreted as “best”. So in the case of some traits, like anthesis date, where a breeder would probably be selecting for germplasm with earlier dates, you can customize Breeding View settings to sort by other traits, change the direction to ascending values, or sort and display the BLUEs. You can also use the custom sort feature of Excel to sort BLUPs within the provided spreadsheet (.xls). Summary of Traits by BLUP Tlaltizapan Summary of Traits: The minimum, mean, maximum, and heritability for each trait based on BLUPs Estimated Genetic Correlations BLUPs Principle Components Biplot Tlaltizapan Principle Components Biplot of BLUPs: Almost 80% of the variation observed at this location is explained by the first two principle components. The first principle component describes 62.42% of the variation, and is correlated most strongly to the traits: field weight (FW), grain yield (GY), plant height (PH), and ear height (EH). The second principle component describes 17.05% of the variation and is correlated most strongly to moisture (MOI). Pairwise Phenotypic Correlations Tlaltizapan Estimated Genetic Correlations: Pairwise comparison of phenotypic traits. There is a strong positive correlation (0.9978) between the related yield measures, grain yield (GY) and field weight (FW). These two yield traits are moderately negatively correlated (-0.6890 & -0.6830 respectively) to anthesis date. In other words, late anthesis is correlated to low yield. Summary Statistics for Individual Trait Raw Data Tlaltizapan Summary Statistics: Anthesis Date (AD) based on raw data Estimated Heritability of Individual Trait Estimated Heritability: Anthesis date (AD) calculated at Tlaltizapan Best Genotypes by Trait Sorted by BLUEs 20 “Best” Anthesis Date (AD) Genotypes Calculated at Tlaltizapan: Genotypes are sorted by BLUEs in descending order by value. For traits, like anthesis date, where small phenotypic measures would be considered “best” we recommend using the custom sort features of Breeding View under the Generate Report node or using Excel to sort BLUEs within the provided spreadsheet (.xls). Standard Errors of Difference Diagnostic Bioplots of Individual Traits Diagnostic biplots illustrate the fit of the trait data to the analysis model based on the residuals. Residuals are estimates of experimental error obtained by subtracting the observed phenotypic measurement from the means predicted by the model. When residuals appear to behave randomly, it suggests that the model fits the data well. On the other hand, if non-random structure is evident in the residuals, it suggests that the model fits the data poorly (NIST/SEMATECH eHandbook of Statistical Methods). Well-Fitted Characteristics of Each Diagnostic Plot Histogram of Residuals: Gaussian, or normal, distribution Fitted-Value Plot: Random distribution, “shot-gun” pattern Normal Plot: Distribution in a straight line across the diagonal Half-Normal Plot: Distribution in a straight line across the diagonal Diagnostic plots for Anthesis Data at Tlaltizapan: Example of a trait exhibiting a good model fit Diagnostic plots for Anthesis Data at Jutiapa: Example of a trait exhibiting a poor model fit References Cullis, B. R., Smith, A. B., & Coombes, N. E. (2006). On the design of early generation variety trials with correlated data. Journal of Agricultural, Biological, and Environmental Statistics, 11(4), 381-393. Li, J., & Ji, L. (2005). Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity, 95(3), 221-227. Murray, D. Payne, R,, & Zhang, Z. (2014) Breeding View, a Visual Tool for Running Analytical Pipelines: User Guide. VSN International Ltd. (.pdf) (associated sample data .zip) NIST/SEMATECH e-Handbook of Statistical Methods, http://www.itl.nist.gov/div898/handbook/ November 2014 Funding & Acknowledgements The Integrated Breeding Platform (IBP) is jointly funded by: the Bill and Melinda Gates Foundation, the European Commission, United Kingdom's Department for International Development, CGIAR, the Swiss Agency for Development and Cooperation, and the CGIAR Fund Council. Coordinated by the Generation Challenge Program the Integrated Breeding Platform represents a diverse group of partners; including CGIAR Centers, national agricultural research institutes, and universities. The statistical algorithms in the Breeding View were developed by VSNInternational Ltd in collaboration with the Biometris group at University of Wageningen. Maize demonstration data was provided by Mike Olsen from the CIMMYT, the International Center for Maize and Wheat Improvement, breeding program. These data have been adapted for training purposes. Any misrepresentation of the raw breeding data is the solely the responsibility of the IBP. This work is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License. ,