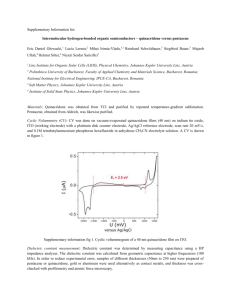
Supplementary Information Description of Image Processing To
advertisement

Supplementary Information Description of Image Processing To determine the number of cells in each image, we used the watershed method.7 A step-bystep depiction of image outputs generated with this method is displayed in Supplementary Figure 2. This method has the ability to distinguish different cells that are clumped together in the same cluster. There are four steps to apply this method. First, the original fluorescence image (Fig. S2A) is converted to grayscale. Since images of cellular fluorescence collected with confocal microscopy have low contrast regions in the middle of images, at top layers, and outside the spheroid at lower layers, all the images are enhanced using a gamma transformation (Fig. S2B). The image is then converted into a binary image in Matlab with the native function ‘graythresh’ (Fig. S2C). The function applies Otsu’s method to find the global threshold level of each image. Any pixels with a higher intensity value than the threshold level will be assigned a value of 1, and those below the threshold are assigned 0. This binary image is an applied open morphology operation to increase the size of cell clusters to define the boundary of all existing cells. The kernel is chosen so that the size of cluster does not increase to the point where significant noise will be introduced. Any holes inside of clusters, which have 0 pixel intensity, are converted into 1 using the ‘imfill’ function to create a completely connected cluster (Fig. S2D). The third step of the program is to define mask and marker images. A mask image is created from the original image to identify possible positions at which there might be cells (Fig. S2E). The marker image allows the program to determine each individual cell by marking locations of higher pixel intensity than the surrounding pixels. To make cells more distinguishable, we applied an adaptive histogram equalization method on a filtered image using a Gaussian filter. Because adaptive histogram equalization increases image contrast corresponding to local areas of image, the resulting image has more detail. Therefore, it decreases the probability that cell clusters will be improperly counted as a single cell. The marker image is created by using the function ‘imextendedmax’ (Fig. S2F). This function finds the regional maxima of the image, and assigns these pixels a value of 1 and all other pixels a value of 0. Finally, after the program successfully compiles the mask image and maker image (Fig. S2G), the image is segmented using the native watershed function in Matlab (Fig. S2H). The number of cells in each 3D stack of images is counted after the segmentation process completes. This program was validated by comparing the numerical output of cells in a random selection of single slice images against hand counts (n = 10 images, 250 cells counted). Using this technique, the program accuracy was determined to be 96.8%. Captions to the Supplementary Figures and Videos Supplementary Fig. 1: Hydrolysis of hydrogels formed at 2.5 w/v% HAS/Gel-S crosslinked with PEGDA or PEGDVS solutions. Gels were formed in solutions that ranged from pH 5 (left) to pH 10 (right) in one-unit increments and were immersed in media. At 14 days, the pH 9 and pH 10 samples crosslinked with PEGDA had hydrolyzed, and at 16 days the remainder of the samples had degraded. Hydrogels crosslinked with PEGDVS did not experience hydrolysis for up to 30 days regardless of pH. Tube labels indicate crosslinker and pH of formulation solution. Supplementary Fig. 2: Step-by-step example image outputs of the developed image processing method. (A) The original image is processed into a (B)grayscale image following gamma transformation, (C) a binary image, (D)and a cluster filled binary image. (E) The mask image, (F) and the marker image are defined and (G) overlaid. (F) Finally, the image is segmented using a watershed transformation (scale bar = 200 μm). Supplementary Videos 1-4: 3D renderings of z-stack images of U118 invasion at days 1, 5, 10, and 14 (videos 1, 2, 3, and 4, respectively). These renderings are composed of the same images displayed in Fig. 4 for the 5 kPa hydrogel with HGF supplementation.