A novel mitochondrial membrane protein, OhmM, limits
advertisement

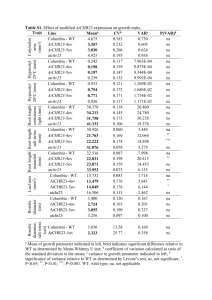
A novel mitochondrial membrane protein, Ohmm, limits fungal oxidative stress resistance and virulence in the insect fungal pathogen Beauveria bassiana Zhangjiang He2, Suhong Zhang2, Nemat O. Keyhani3, Yulin Song2, Shuaishuai Huang2, Yan Pei2, Yongjun Zhang1, 2* Supporting Information Figure S1. Molecular manipulations of Ohmm; targeted gene disruption, complementation, and overexpression. (A) Ohmm locus and gene replacement vector p∆Ohmm. p∆Ohmm contains bar cassette flanked by 3' and 5' border sequences of Ohmm. Homologous recombination (cross over event marked by “X”) resulting in a region of Ohmm was replaced by the bar cassette. Arrow indicates the orientation of transcription in the ORF. (B) Ohmm complementation vector; pCB-Ohmm, containing the sulfonylurea resistance marker (sur). (C) Ohmm overexpression vector. The vector contains the Ohmm-coding region under the control of the fungal constitutive promoter PgpdA from Aspergillus nidulans. (D) PCR analysis of wild-type strain (WT), ΔOhmm, p∆Ohmm ectopic integrant (Control), and Ohmm::Ohmm transformant (complemented mutant). Desired integration events were confirmed by PCR using primers S1 and S2. (E) RT-PCR confirmation of loss of gene expression in the ΔOhmm disruption mutant and recovery in the complemented strain (ΔOhmm::ΔOhmm). Total RNA was isolated as described in the Methods section. Expression of Ohmm was examined using actin as the reference gene in wild type (WT), ΔOhmm, p∆Ohmm ectopic integrant, and ΔOhmm::Ohmm transformants. (F) Real-time RT-PCR determination of Ohmm transcript levels in the wild type (WT) and Ohmm overexpression (OE-Ohmm) strains; actin and β-tubulin were used as reference genes. Error bars indicate standard deviations. (G) Southern blot analysis of p∆Ohmm ectopic integration transformant (control), ΔOhmm, and Ohmm overexpression (OE-Ohmm) strains. Genomic DNAs were digested with EcoRI and probed using the bar gene as described in the Methods. Figure S2. Gene disruption of Ohmm in the Hog1 strain. (A) Ohmm locus in the ΔHog1 strain and gene replacement vector p∆Ohmm. The p∆Ohmm contains the sur cassette flanked by border sequence of Ohmm. Homologous recombination (cross over event marked by “X”) resulting in a region of Ohmm replaced by the sur cassette. Arrow indicates the orientation of transcription in the ORF. (B) PCR analysis of ΔHog1 strain and the double ΔHog1ΔOhmm mutan. The desired integration event was confirmed by PCR using primers S1 and S2 and the integrity of the ΔHog1 strain was verified by PCR using HS1 and HS2 (4861 bp, Hog1 homologous recombination cassette containing the bar resistance marker). (C) RT-PCR confirmation of loss of Ohmm gene expression in the ΔHog1ΔOhmm mutant. Total RNA was isolated as described in the Methods section. Expression of Ohmm was examined using actin as the reference gene in Bbhog1 mutant (Δhog1) and the double Hog1Ohmm mutant strain. (D) Southern blot analysis of the double Hog1Ohmm mutant. Genomic DNAs were digested with EcoRI and probed using partial fragment of the sur gene as described in the Methods. Table S1. mRNA size and DNA sequence analysis of SSH cDNA clones expressed in the WT strain but with markedly decreased expression in the Hog1 mutant under osmotic stress for 24 h Clone (GenBank accession No.) Size (bp) E value H64 226 9.00E-42 H77 251 H231 Identity (%) Accession no. Homologous sequence in GenBank 97 EJP66004 Beauveria bassiana AMP-binding enzyme 1.00E-08 92 XP_002413831 Ixodes scapularis proteasome subunit alpha type 516 5.00E-36 100 EJP63790 Beauveria bassiana aldo/keto reductase H388 435 8.00E-75 97 EJP66797 Beauveria bassiana short-chain dehydrogenase H55 511 7.00E-21 43 XP_003855870 Zymoseptoria tritici hypothetical protein H59 553 5.3 30 ZP_09618764 H128 594 8.00E-53 89 EJP68629 H209 309 2.00E-62 100 EJP66466 H669 768 1.00E-50 86 EJP66533 H83 280 3.00E-15 97 EJP64424 H350 279 2.00E-07 100 EJP69867 H232 413 unknown H235 205 unknown H464 206 unknown Legionella drancourtii hypothetical protein LDG_5091 Beauveria bassiana hypothetical protein BBA_02631 Beauveria bassiana hypothetical protein BBA_04406 Beauveria bassiana hypothetical protein BBA_04473 Beauveria bassiana hypothetical protein BBA_06418 Beauveria bassiana hypothetical protein BBA_00736 Table S2. Oligonucleotides used in this study Paired primers Sequence (5'–3')* Remarks YADE-based PCR walking Y5P1/Y5P2 AGACTGTGCTCAACGCCATG/ AGGACCTGGGCATGATTCTC Cloning 5' fragment of Ohmm Y3P1/Y3P2 ATGGCGTTGAGCACAGTCTT/ATGAACCGGTGCGACTGCTC Cloning 3' fragment of Ohmm Cloning cDNA of Ohmm using 3' RACE RaF1 CATGATGGCGACCTCTATCA RaF2 TAATCCTTGGACGCTGCGAC RaR1 TTAGCTATCCCAAATAGCT RaR1 CAAGAGCCTTCTTGTCCGTC Cloning FAD/NAD(P)-binding region FD1/FD2 CGGAATTCCGATGGCGGCCGTTCTCTCCTT /ATAAGAATGCGGCCGCTAAACTATTTAGCTATCCCAAATAGCTCCAT GFP::Ohmm fusion vector GF1/GF2 CGGAATTCCGATGGTGAGCAAGGGCGAGGA/ GACTTGTACAGCTCGTCCAT Cloning of eGFP OG1/OG2 ATGGACGAGCTGTACAAGTGGCTCGGGCATGATGGCGACCTCTATCAT/ GCGATATCCTATTAGCTATCCCAAATAGCT Cloning of Ohmm containing upstream sequence Ohmm disruption vector p∆Ohmm construction and screen or confirmation of disruption strain L1/L2 CCCAAGCTTATCATGATGGCGACCTCTAT /TCAATGTCATCTTCTGTCGACATGACGAGGGTGCCACCATT Cloning of 5'-end of Ohmm R1/R2 TGCCCGTCACCGAGATCTAATAGTCAATACCATGAGCAGACAT /GCTCTAGACGTTGAGAAGCACCACCACGGA Cloning of 3'-end of Ohmm B1/B2 GTCGACAGAAGATGACATTGA/CTATTAGATCTCGGTGACGGGCA Cloning of bar cassette S1/S2 ATCTCTCTCAAGATTCTCAT/TACACATTGTTATTAGGAGT Screen or confirmation disruption strain of Cloning Ohmm sequence used for construction of reverse complement vector pCB-Ohmm RC1/RC2 CCTACGTATACAGCTGCAATGATTGGCT /GCTCTAGATCCTTGTCTGGTAGGCGTAG Cloning Ohmm coding region used for construction of overexpression vector O1/O2 GGAATTCAAGACGTATATATGGTGCGA /GCGATATCATTTAGCTATCCCAAATAGCT Construction of Ohmm disruption vector used for disruption of the gene in ∆Hog1 mutant L3/L4 R3/R2 SuF/SuR GGAATTCATCATGATGGCGACCTCTAT /TCAATGTCATCTTCTGTCGACATGACGAGGGTGCCACCATT Cloning of 5'-end of Ohmm CGGGAATTGCATGCTCTCACTCAATACCATGAGCAGACAT /GACTAGTCGTTGAGAAGCACCACCACGGA Cloning of 3'-end of Ohmm GTCGACAGAAGATGACATTGA/GTGAGAGCATGCAATTCCCG Cloning sur cassette Confirmation of Hog1 disruption strain (∆Hog1) HS1/HS2 GAATTCATTCACCCTTTGCGT/AAGTAATACCCTTTCAGAGCA RT-PCR and real-time RT-PCR aF/aR βtF/βtR TTGGTGCGAAACTTCAGCGTCTAGTC /TCCAGCAAATGTGGATCTCCAAGCAG TACTCTACGATTCGTCAAGT/TGCTGGAACAGAGCCGTCTT ohF/ohR ATCGCGACGCTCTTTAACCG/GCTATCCCAAATAGCTCCAT FluG1/2 CCTCCCTAGTTTGGTCGCTTTCTC / CGCTGTCGGTAATCTGCTCCTC FlbA1/2 CCAATCCACTCGCCGCTCTC / CGGAGGAAAGAGAATCGGTAGAGG RT-PCR and qRT-PCR analysis of actin RT-PCR and qRT-PCR analysis of β-tubulin RT-PCR and qRT-PCR analysis of BbohmM RT-PCR and qRT-PCR analysis of FluG RT-PCR and qRT-PCR analysis of FlbA FlbB1/2 GCACTGACACGCCGACAAGAGC / CCGCCGCCGAAGCCTGTTG FlbC1/2 TCCATCTCCAACTTGCTGGGTCTC / GGCGGCGTAGGCGGAAGG FlbD1/2 CGGCAAGCGATGGGCAGAGATTG /ACGAGCAAGGTGACGGTAGAGGTG hyd1F/R ATCTACTGCTGCAACGAGAA/TACTGGATAAGACTGCCAAT hyd2 F/R AGTGTCAAGACTGGCGACAT/ATCCGAGGACGGTGATGGGA RT-PCR and qRT-PCR analysis of hyd2 CatB-F/R AAGGCCATTCCCGTCATCAT/ AGCTCGCGGTCCCAGCATCT RT-PCR and qRT-PCR analysis of CatB CatA-F/R AGAGCAACGGCAAGCGTGTA / AGAAGTCGCTGCTCAGGGTA CatC-F/R CatP-F/R GAGGAGCCCAGCAACGCACAAGAG/ CTGAGGACGACAAGGCCGCCATTC GCTGGGCTGATCTGCTGGTCCTTG / TCCTTGCTGTAACGGTGGCTGTCG CatD-F/R CGGCTGCGGTGTCTTGTCCATAC / CCTTGTCGGCGTTCTGGCGAAG Sod1-F/R TCCACATCCACACCTTTGGT / AGGTCCAGCGTTGCCAGTCT Sod2-F/R AGCTCCTCGCCGCCATCACCA / AGCCGTCTTCCAGTTGATGA Sod3-F/R ATGACGCCAAGGCTGCTGCT / ATCAATGCCCAGTAGAGGCT Sod4-F/R CGAGATGGTCCTTACGGCTTCAG / GCTCCCAGGTGTTGAGGCATAG HPX1-F/R GCCGCCGCTTCTACTCGTCTG / CGTTGCTGCCGCCAGTACCG Trx1-F/R GCCATTTCGCCCTTCTTCACCAAG / CGCAGCCGCCGTTGTTGTAG RT-PCR and qRT-PCR analysis of CatA RT-PCR and qRT-PCR analysis of CatC RT-PCR and qRT-PCR analysis of CatP RT-PCR and qRT-PCR analysis of CatD RT-PCR and qRT-PCR analysis of Sod1 RT-PCR and qRT-PCR analysis of Sod2 RT-PCR and qRT-PCR analysis of Sod3 RT-PCR and qRT-PCR analysis of Sod4 RT-PCR and qRT-PCR analysis of HPX1 RT-PCR and qRT-PCR analysis of trx1 *The enzyme sites are italicized. RT-PCR and qRT-PCR analysis of FlbB RT-PCR and qRT-PCR analysis of FlbC RT-PCR and qRT-PCR analysis of FlbD RT-PCR and qRT-PCR analysis of hyd1