Section 13
advertisement

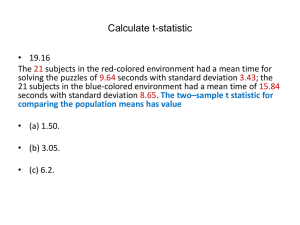
STAT 405 - BIOSTATISTICS
Handout 13 – More on Chi-Square Tests
This handout covers material found in Section 10.6 of your text (plus some additional material).
THE CHI-SQUARE TEST OF INDEPENDENCE
Previously, we discussed the chi-square test for comparing two proportions. All of these
examples involved data summarized in the form of a 2x2 contingency table. Frequently,
however, one or both variables of interest have more than two categories. The chi-square test
can also be used to analyze these data.
EXAMPLE: The following table classifies a sample of psychiatric patients by their diagnosis
and by whether their treatment prescribed drugs.
Diagnosis
Drugs No Drugs
Schizophrenia
105
8
Affective Disorder
12
2
Neurosis
18
19
Personality Disorder
47
52
Special Symptoms
0
13
Pearson Chi-Square Statistic
Again, the traditional test statistic for analyzing these data is given as follows:
Test Statistic =
i
(Observedi - Expectedi ) 2
Expectedi
To calculate this test statistic, we need to first find the counts that would be EXPECTED if there
were no association between the two variables.
The Observed Counts
Diagnosis
Drugs No Drugs
Schizophrenia
105
8
Affective Disorder
12
2
Neurosis
18
19
Personality Disorder
47
52
Special Symptoms
0
13
The Expected Counts
Diagnosis
Schizophrenia
Affective Disorder
Neurosis
Personality
Disorder
Special Symptoms
Total
Drugs No
Drugs
Total
113
14
37
99
182
94
13
276
1
Once again, we can use SAS to calculate the expected counts and also the test statistic:
data psych;
input diagnosis$ drug$ count;
datalines;
schizophrenia
yes 105
schizophrenia
no
8
affective_disorder
yes 12
affective_disorder
no
2
neurosis
yes 18
neurosis
no
19
personality_disorder
yes 47
personality_disorder
no
52
special_symptoms
yes 0
special_symptoms
no 13
;
proc freq order=data;
tables diagnosis*drug / all expected nopercent nocol norow;
weight count;
run;
2
The test can be carried out as follows:
Ho: There is no association between diagnosis and drug prescription.
Ha: There is an association between diagnosis and drug prescription.
p-value =
Conclusion:
Likelihood-Ratio Statistic
An alternative statistic results from the likelihood-ratio method for significance tests. The
general form of this statistic is given as follows:
G2 2
Observ edi
(Observ ed ) ln Expected
i
i
i
When the null hypothesis is true, this test statistic approximately follows the chi-square
distribution with df = (r-1)(c-1).
Questions:
1. What is the minimum value of G2? When does the statistic assume this value?
2. What do large values of G2 indicate? Explain.
3. Verify the value of G2 from the SAS output:
3
G2 2
Observ edi )
i
(Observ ed ) log Expected
i
i
4. Does your conclusion change when using the likelihood ratio statistic versus the Pearson
chi-square statistic? Explain.
4
Cell Chi-square
In our example, we have found evidence for an association between diagnosis and drug
prescription. To better understand the nature of this association, we can examine the difference
between the observed and expected counts in each cell. However, for cells that have larger
expected frequencies, a larger difference tends to exist between the observed and expected
counts; therefore, examining the raw difference between the two is insufficient. Instead, we
examine adjusted residuals:
Observed - Expected
Expected(1 - π̂ i )(1 - π̂ j )
,
where π̂i and π̂ j represent the row and column totals of the joint probabilities, respectively.
For example, we can calculate the adjusted residuals for each cell in our contingency table:
5
When the null hypothesis is true, each adjusted residual has a large-sample standard normal
distribution. Therefore, an adjusted residual that exceeds about 2 or 3 in absolute value
indicates a lack of fit of the null hypothesis in that cell.
Questions:
1. What do you learn from the adjusted residuals in our example? Explain.
2. Do your conclusions agree with what you see in the mosaic plot?
3. Do your conclusions agree with what you learn from each cell’s contribution to the
Pearson chi-square statistic? Explain.
proc freq order=data;
tables diagnosis*drug / cellchi2 nopercent nocol norow;
weight count;
run;
6
THE CHI-SQUARE TEST FOR TREND IN BINOMIAL PROPORTIONS
One form of this test discussed in detail in Section 10.6 of your text. This handout will present
an alternative method from An Introduction to Categorical Data Analysis by Alan Agresti.
EXAMPLE: The data in the following table refer to a prospective study of maternal drinking
and congenital malformations. After the first three months of pregnancy, the women in the
sample completed a questionnaire about alcohol consumption. Following childbirth,
observations were recorded on presence or absence of congenital sex organ malformations.
Malformation
Alcohol Consumption Absent Present
0
17,066
48
<1
14,464
38
1-2
788
5
3-5
126
1
6+
37
1
We could use a chi-square test of independence to analyze these data (although the test may not
be valid because of too many small expected cell counts).
However, even if we were able to use this test to show that some relationship exists between
alcohol consumption and congenital malformations, the results would not tell us specifically
about the nature of the relationship. In particular, we may want to provide evidence for an
increasing trend in the proportion with a congenital malformation in each succeeding row.
To do this, we must first introduce a score variable, Si, corresponding to the ith group. This
variable can represent some particular numeric attribute of the group; however, in many cases
for simplicity, 1 is assigned to the first group, 2 to the second, etc. These scores should have the
same ordering as the category levels, and they should reflect the distances between categories
(with greater distances between categories treated as farther apart). A few different options are
shown below.
Score, Option 1
(Midpoint of Category)
Score, Option 2
(For Simplicity)
0
0
<1
.5
1-2
1.5
3-5
4
6+
7
1
2
3
4
5
7
The chi-square test for trend can be carried out as follows:
Set up the hypotheses:
Ho: There is no trend among the proportions (independence).
Ha: The proportions are an increasing or decreasing function of the scores (non-zero
correlation).
Calculate the test statistic:
The test statistic is computed as χ 2 (n 1)r 2 , where
r = the Pearson correlation between the two variables
n = total number of subjects in the study
Under the null hypothesis and for large samples, this test statistic approximately follows a chisquare distribution with df =1.
The correlation can be computed using SAS PROC CORR:
data a;
input consumption malformation count;
datalines;
0
0
17066
.5
0
14464
1.5 0
788
4
0
126
7
0
37
0
1
48
.5
1
38
1.5 1
5
4
1
1
7
1
1
;
proc corr;
var consumption malformation;
weight count; run;
χ 2 (n 1)r2 =
8
Find the p-value:
The approximate p-value is given by the area to the right of the test statistic under the chisquare distribution with df = 1.
data ChiSquareprob;
Prob=1-CDF('ChiSquare',6.57,1); output;
proc print;
run;
This test can also be carried out using SAS PROC FREQ:
proc freq order=data;
tables Consumption*Malformation / cmh1;
weight count;
run;
9
Test for Trend Compared to Pearson’s Chi-Square and Likelihood Ratio Tests
There are a few reasons why one would want to use the test for trend over the more traditional
chi-square methods.
When the association truly has either a positive or negative trend, the ordinal test (i.e.,
test for trend) is more powerful than Pearson’s chi-square or the likelihood ratio test.
For small to moderate sample sizes, the chi-square approximation is likely to be worse
for Pearson’s chi-square or the likelihood ratio test statistic than it is for the trend test
statistic.
Choice of Scores
For most data sets, the choice of scores has little effect on the results. However, in some
instances, this may not be the case. For example, consider using our second option for scores:
data a;
input consumption malformation count;
datalines;
1
0
17066
2
0
14464
3
0
788
4
0
126
5
0
37
1
1
48
2
1
38
3
1
5
4
1
1
5
1
1
;
proc freq order=data;
tables Consumption*Malformation / cmh1;
weight count;
run;
10
The Midrank Approach
An alternative approach uses the data to form scores automatically. These are called midranks
and are calculated as follows for our data:
Alcohol Consumption Absent Present Total Midrank
0
17,066
48
17,114
8557.5
<1
14,464
38
14,502 24,365.5
1-2
788
5
793
32,013.0
3-5
126
1
127
32,473.0
6+
37
1
38
32,555.5
For example, the 17,114 subjects who consume no alcohol share ranks 1 through 17,114. We
assign each of them to the average of these ranks: (1+17,114)/2 = 8557.5.
The 14,502 subjects who consume one or fewer share ranks 17,115 through (17,114+14,502) =
31,616. This gives a midrank of (17,115 + 31,616)/2 = 24,365.5.
This test can be carried out in SAS as follows:
data a;
input consumption malformation count;
datalines;
1
0
17066
2
0
14464
3
0
788
4
0
126
5
0
37
1
1
48
2
1
38
3
1
5
4
1
1
5
1
1
;
proc freq order=data;
tables Consumption*Malformation / cmh1 scores=ridit;
weight count;
run;
11
Questions:
1. What is our conclusion?
2. Why is this result so different from the results obtained using other scores?
3. Which scoring method do you think should be used? Why?
12
Chi-Square Test for Trend in Binomial Proportions in (2 x k) Tables
For test details and example see pages 430-437 in your text. The test itself is certainly doable by
hand, but why? Learning to program R is a worthwhile endeavor as it allows you to program
non-standard test procedures yourself.
Below is some rudimentary R code for performing the test. It takes three vectors of input, the
frequencies associated with the “cases” (xi), the total number of observations in each of the
levels of the score variable (ni), and the levels of the score variable S (Si), which by default is
1,…,# of levels of the score variable.
Ptrend = function(x,n,scores=1:length(x)) {
# The next seven lines of code perform all of the required
# computations.
A = sum(x*scores) - sum(x)*sum(n*scores)/sum(n)
pbar = sum(x)/sum(n)
qbar = 1 - pbar
nsum = sum(n)
B = pbar*qbar*(sum(n*scores^2)-(1/nsum)*sum(n*scores)^2)
X2 = (A^2)/B
pval = 1 - pchisq(X2,1)
# The remainder of the code makes the output look pretty and constructs # two
plots to visualize the results.
cat("\n")
cat("Test for Trend in Binomial Proportions\n")
cat("=======================================================\n")
cat(paste("A =",format(A,dig=6),"\n"))
cat(paste("Chi-square Statistic =",format(X2,dig=6),"\n"))
cat(paste("p-value =",format(pval,dig=6),"\n"))
phat = x/n
# Code to construct the plots
par(mfrow=c(1,2),pty="s")
plot(scores,phat,xlab="Scores",ylab="Sample Proportion (p-hat)",main="p-hat
vs. Scores",cex=.4,pch="o")
datamat = cbind(x,n-x)
colnames(datamat) = c("Y","N")
rownames(datamat) = as.character(scores)
mosaicplot(datamat,col=4:5,main=”p Trend Test”)
par(mfrow=c(1,1),pty="m")
}
An example is given on the next page.
13
Example: Breast Cancer and Age at 1st Birth (see pg. 433)
>
>
>
>
x <- c(320,1206,1011,463,220)
n <- c(1742,5638,3904,1555,626)
scores <- c(1,2,3,4,5)
Ptrend(x,n,scores)
Test for Trend in Binomial Proportions
=======================================================
A = 567.16
Chi-square Statistic = 129.012
p-value = 0
14