Chi-Square Analysis in Genetics: Worksheet
advertisement
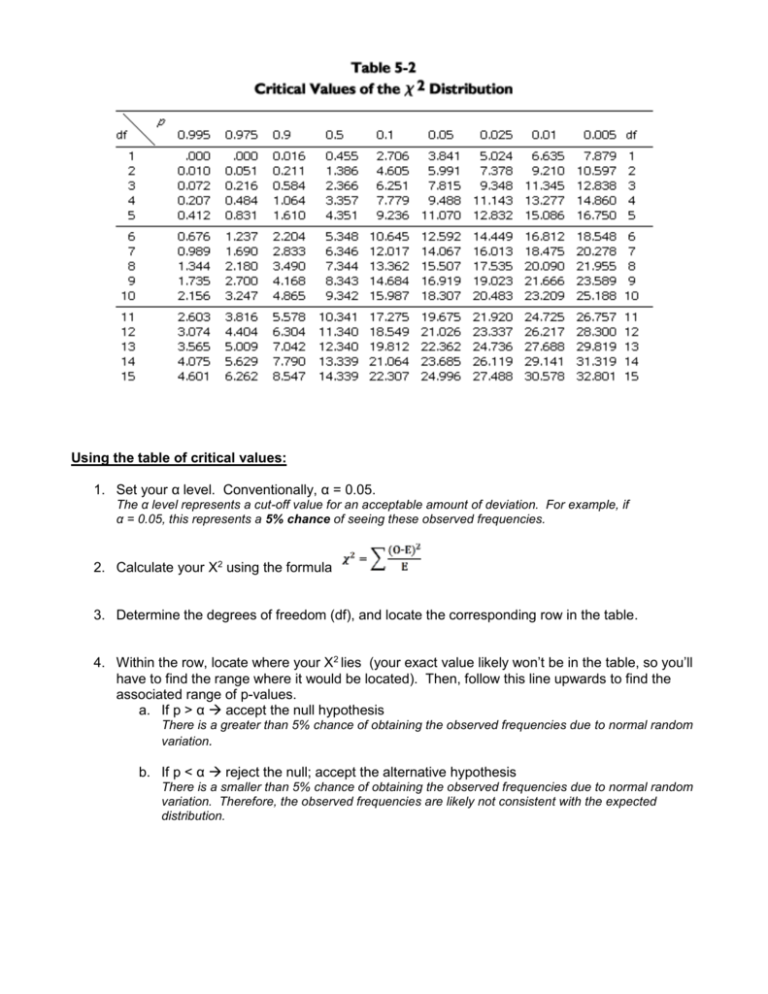
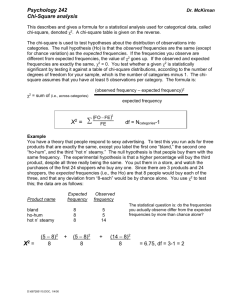
Using the table of critical values: 1. Set your α level. Conventionally, α = 0.05. The α level represents a cut-off value for an acceptable amount of deviation. For example, if α = 0.05, this represents a 5% chance of seeing these observed frequencies. 2. Calculate your Χ2 using the formula 3. Determine the degrees of freedom (df), and locate the corresponding row in the table. 4. Within the row, locate where your Χ2 lies (your exact value likely won’t be in the table, so you’ll have to find the range where it would be located). Then, follow this line upwards to find the associated range of p-values. a. If p > α accept the null hypothesis There is a greater than 5% chance of obtaining the observed frequencies due to normal random variation. b. If p < α reject the null; accept the alternative hypothesis There is a smaller than 5% chance of obtaining the observed frequencies due to normal random variation. Therefore, the observed frequencies are likely not consistent with the expected distribution. Applying Chi-Square Analysis to Genetics Problems So much of Mendelian genetics involves ratios. See the table below for the characteristic ratios we have seen. Table 3. Characteristic phenotypic and genotypic ratios associated with monohybrid and dihybrid crosses. F2 ratio Phenotypic Genotypic Monohybrid cross 3:1 1:2:1 Dihybrid cross 9:3:3:1 ------We can use the chi-square test to compare observed frequencies of phenotypic and genotypic classes to the frequencies that would be expected, based on Mendelian ratios. Recall that these ratios are based on characters that assort independently (are not linked). One application of the chi-square test to genetics can therefore be to indicate whether two characters are linked or not. Example 1 In flowers, red petals (A) are dominant to white petals (a). You think that a particular plant is heterozygous (Aa), and to test this theory you perform a test cross. You obtain 120 progeny, 55 of which are red, and 65 of which are white. a) In this test cross, what is the genotype of the second plant? aa b) What are the expected proportions of red to white plants in the progeny? Red: 50% c) White: 50% What are the expected frequencies of red to white plants? Red: 50% x 120 = 60 White: 60 d) Define a null hypothesis and an alternative hypothesis: H0: The observed data follows a 1:1 distribution HA: The observed data does not follow a 1:1 distribution e) Define a significance level: α = _0.05___ f) Fill in the chi-square calculation table: Class O E (O-E)2 red 55 60 25 white 65 60 25 Total = g) (O-E)2/E 0.417 0.417 X2 = 0.834 How many degrees of freedom are in your data? _1_________ h) Compare your test value, X2, to the table of critical values. What is the P value for your X2? __0.1-0.5 (or 10%-50%)_____________ i) Do you reject or accept the null hypothesis, H0? j) Draw a general conclusion based on your test: □ accept □ reject □ The observed frequencies match the expected distribution □ The observed frequencies do not match the expected distribution k) Draw a specific conclusion based on your test: □ The red plant is heterozygous Aa □ The red plant is homozygous AA Example 2. A plant geneticist has two pure lines, one with purple petals, and one with blue. She hypothesizes that the phenotypic difference is due to two alleles of one gene. To test this idea, she aims to look for a 3:1 ratio in the F 2. She crosses the lines and finds that all F1 progeny are purple. The F1 plants are selfed, and 400 F2 plants are obtained. Of these, 320 are purple and 80 are blue. Use the chi-square test to determine if these results fit her hypothesis. α= __0.05_____ H0: The observed data follows a 3:1 distribution HA: The observed data does not follow a 3:1 distribution Class O E (O-E)2 (O-E)2/E Purple 320 300 400 1.33 80 100 400 4.00 400 400 Blue df= _1______ Total = X2= 5.33 P value: 0.01-0.025________ reject the null hypothesis accept the alternative hypothesis General Conclusion: The data does not follow a 3:1 distribution Specific Conclusion: Flower colour is not due to two alleles of one gene Example 3. Thomas Hunt Morgan studied the inheritance of characters in fruit flies, Drosophila melanogaster. He mated fruit flies with the following phenotypes/genotypes: Parent Phenotype Gray body/Normal wings Genotype Gg Ww Black body/Vestigial wings gg ww He obtained 2300 progeny with the following genotypes and frequencies: Gg Ww 965 Gg ww 206 gg ww 944 gg Ww 185 Use the chi-square test to determine whether these results support the hypothesis that the genes for body colour and wing type assort independently. α= 0.05 H0: The observed data follows a 1:1:1:1 distribution (based on Punnett square prediction) HA: The observed data doesn’t follow a 1:1:1:1 Class O E (O-E)2 (O-E)2/E GgWw 965 575 152100 264.522 ggww 944 575 136161 236.802 Ggww 206 575 136161 236.802 ggWw 185 575 152100 264.522 Total = X2= 1002.648 2300 df= 3 P value: <<0.005 reject the null hypothesis accept the alternative hypothesis General Conclusion: The observed data doesn’t follow a 1:1:1:1 Specific Conclusion: The genes for body colour and wing type do not assort independently